Exome sequencing in severe non-syndromic specific learning and language disorders in a French cohort
- PMID: 41168819
- PMCID: PMC12574064
- DOI: 10.1186/s13229-025-00688-8
Exome sequencing in severe non-syndromic specific learning and language disorders in a French cohort
Abstract
Background: Specific learning disorders (SLDs) affect approximately 5% of school-age children. In France, genetic investigations of complex non-syndromic SLD cases include chromosomal microarray analysis and fragile X syndrome testing. However, the examples of genes being described in intellectual disability or autism spectrum disorder and also reported in patients with complex and severe SLDs are multiplying. International efforts using exome sequencing have identified monogenic diseases that explain severe SLDs in some instances. The aim of our study was to investigate the value of exome sequencing in children with SLDs without intellectual disability and autism spectrum disorder.
Methods: We initiated a prospective study using exome sequencing in patients with well-documented, severe SLD.
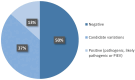
Results: Analysis of 82 patients revealed pathogenic/likely pathogenic variants in 11 (13.4%) patients (ADNP, BRAF, CREBBP, KCNN2, KIF1A, RERE, SCN8A, SET, SMARCC2 (x2), TRIO). In addition, 38 variants of uncertain significance in candidate genes for severe neurodevelopmental disorders (NDDs) or in genes of unknown significance that could contribute to the phenotype were identified in 30 patients.
Limitations: The study of 82 patients does not provide sufficient statistical power to conclude that exome testing adds value over chromosomal microarray analysis, or to determine which patient profiles are more likely to benefit from genetic testing. Studies with larger patient numbers are needed to increase statistical power.
Conclusions: This study confirms the involvement of NDD genes in milder phenotypes and suggests the potential of exome sequencing for diagnosing severe SLDs. Further research on larger samples is required to determine which SLDs are most likely to benefit from pangenomic explorations and to clarify the implication of candidate variants.
Keywords: Developmental language disorders; Exome sequencing; Neurodevelopmental disorders; Specific learning disorders.
Conflict of interest statement
Declarations. Ethical approval and consent to participate / consent for publication: Written informed consent was obtained from individuals and legal guardians/next of kin of minors for publication of data included in this article. The two patients whose images are presented in this article consented to the photographs being published. Molecular studies with informed consent were conducted in the framework of the DISCOVERY project (2016-A01347-44). The samples were part of the GAD collection DC2011-1332. This study was conducted in accordance with the ethical principles of the Declaration of Helsinki and the International Conference on Harmonisation guideline for good clinical practice. Competing interests: The authors declare no competing interests.
Figures


References
-
- American psychiatric association, editor. Desk reference to the diagnostic criteria from DSM-5. Washington (D.C.): American psychiatric publishing; 2013.
-
- Les troubles du langage et des apprentissages. Ministère du Travail, de la Santé, des Solidarités, des Familles, de l’Autonomie et des Personnes handicapées. https://sante.gouv.fr/prevention-en-sante/sante-des-populations/enfants/.... Accessed 12 Oct 2025.
-
- Nouvelle stratégie nationale pour les troubles du neurodéveloppement: autisme, Dys, TDAH, TDI | handicap.gouv.fr. 2023. https://handicap.gouv.fr/nouvelle-strategie-nationale-pour-les-troubles-.... Accessed 12 Oct 2025.
-
- Finucci JM, Guthrie JT, Childs AL, Abbey H, Childs B. The genetics of specific reading disability. Ann Hum Genet. 1976;40:1–23. 10.1111/j.1469-1809.1976.tb00161.x. - PubMed
-
- Rossi AO. Genetics of learning disabilities. Behav Neuropsychiatry. 1972;4:2–7. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

