Mitochondrial dysfunction in PRRSV-2-infected macrophages
- PMID: 41169353
- PMCID: PMC12568354
- DOI: 10.3389/fimmu.2025.1670488
Mitochondrial dysfunction in PRRSV-2-infected macrophages
Abstract
Introduction: Porcine reproductive and respiratory syndrome virus (PRRSV) is one of the most economically devastating viruses for the global swine industry. PRRSV has a known tropism for lung macrophages, where it causes impaired immune responses. This study evaluated the metabolic and immune profiles of primary porcine alveolar macrophages (PAMs) and pulmonary intravascular macrophages (PIMs) infected with different strains of PRRSV-2 isolated from North Carolina (NC) pig herds (NC134, NC18-9-7 referred to as NC174, and NC20-1 referred to as NC144), and VR2232, a PRRSV-2 prototype strain.
Materials and methods: Primary enriched mononuclear phagocytes were infected in vitro with NC134 and NC174, sorted, and processed. The total RNA was used for a transcriptomic approach; additionally, gene expression was further validated using RT-qPCR and NanoString technology. Complementary functional assays with additional NC strains were used to further investigate the mitochondrial and metabolic dysfunction, as well as the oxidative stress induced by PRRSV-2 infection.
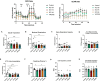
Results: PAMs infected with both NC PRRSV-2 strains NC174 and NC134 showed similar transcriptomic profiles during the early stage of infection, with downregulation of genes involved in the oxidative phosphorylation and electron transport chain pathways. PIMs infected with both NC174 and NC134 strains showed limited alteration in the transcriptomic profiles compared to uninfected cells. Genetic reprogramming matched the PRRSV-2-induced mitochondrial impairment observed in functional assays performed using Seahorse technology. Mitochondrial respiration displayed slightly different profiles between PIMs and PAMs infected with the different PRRSV-2 strains, with PAMs showing a more substantial decrease in mitochondrial fitness compared to control cells. When reactive oxygen species (ROS) and nitric oxide (NO) production were evaluated, no differences were observed between PRRSV-2-infected PAMs and PIMs and control cells.
Conclusion: These results provide valuable insights into the pathogenetic mechanism of different NC PRRSV-2 strains by focusing on the alteration in mitochondrial function in lung macrophages during early infection and highlighting differences in lung macrophage responses to distinct PRRSV-2 strains.
Keywords: NanoString; PRRSV-2; macrophages; mitochondrial dysfunction; pig; seahorse technology; transcriptomics.
Copyright © 2025 Vu Manh, Frias-De-Diego, Williams, Byrne, Sirisereewan, Hicks, Liu and Crisci.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author EC declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




References
-
- Osemeke O, Silva GS, Corzo CA, Kikuti M, Vadnais S, Yue X, et al. Economic impact of productivity losses attributable to porcine reproductive and respiratory syndrome virus in United States pork production, 2016-2020. Prev Vet Med. (2025) 244:106627. doi: 10.1016/j.prevetmed.2025.106627, PMID: - DOI - PubMed
-
- VanderWaal K, Pamornchainavakul N, Kikuti M, Linhares DCL, Trevisan G, Zhang J, et al. Phylogenetic-based methods for fine-scale classification of PRRSV-2 ORF5 sequences: a comparison of their robustness and reproducibility. Front Virol. (2024) 4. doi: 10.3389/fviro.2024.1433931 - DOI
-
- Diseases of Swine, Twelfth Edition . Edited by Jeffrey J. Zimmerman, Eric R. Burrough, Locke A. Karriker, Kent J. Schwartz. © 2026 John Wiley & Sons, Inc. Published 2026 by John Wiley & Sons, Inc.
-
- Zeller MA SA, Sharma A, Trevisan G, Zhang J, Harmon K, Gauger PC. ISU PRRSView 2023 . Available online at: https://prrsv.vdl.iastate.edu/about.php (Accessed July 21, 2025).
MeSH terms
LinkOut - more resources
Full Text Sources

