Cytosine methylase and hydroxymethylase activity in mammalian mitochondria
- PMID: 41170371
- PMCID: PMC12568578
- DOI: 10.3389/fcell.2025.1677402
Cytosine methylase and hydroxymethylase activity in mammalian mitochondria
Abstract
Introduction: Mitochondria are integral components of eukaryotic cells, functioning as energy powerhouses and key mediators of diverse metabolic and signaling cascades. As endosymbiotic remnants, these unique organelles retain and express their own DNA. Mitochondrial DNA (mtDNA) is packaged into DNA-protein complexes called nucleoids, and is also subject to epigenetic modification. We identified a mitochondrial isoform of DNA methyltransferase 1 (mtDNMT1) that binds to mtDNA in critical control regions; however, its enzymatic activity remained unexplored.
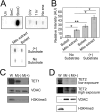
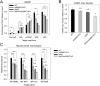
Results: Here, we show that endogenously-tagged mtDNMT1 purified from mitochondria exhibits time- and concentration-dependent CpG-specific DNA methyltransferase activity, but it is not working alone: DNMT3b cooperates with mtDNMT1 to methylate mtDNA and regulate mitochondrial transcription. In addition, we detect ten-eleven translocase (TET)-like hydroxymethylase activity in mitochondria, demonstrating that mechanisms for both writing and erasing 5-methylcytosine marks are functional in this organelle. CRISPR/Cas9-mediated inactivation of mtDNMT1 and/or DNMT3b activity resulted in a stepwise decrease in mitochondrial methylation across the heavy and light strand promoters of mtDNA, with a significant reduction in transcription of several mtDNA-encoded OXPHOS genes. Interestingly, the effects of mtDNA methylation on mitochondrial transcription are diametrically opposed to the role of promoter methylation in the nucleus, suggesting a novel mode of gene regulation in mitochondria. Cells lacking mtDNMT1 and/or DNMT3b also exhibited a modest reduction in mtDNA content, suggesting that methylation impacts both mtDNA transcription and replication.
Discussion: These observations implicate mtDNA methylation in the fine-tuning of mitochondrial function and suggest a role for aberrant mitochondrial methylase activity in disease.
Keywords: DNA demethylation; DNA methylation; DNA methyltransferase; DNA replication; epigenetics; mitochondrial DNA (mtDNA); transcription.
Copyright © 2025 Shock, Thakkar, Robinson and Taylor.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declare that they were a review editor for Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials

