Virulence and mutations analysis based on the whole genome of a Brazilian Corynebacterium diphtheriae strain isolated from a cutaneous infection
- PMID: 41170434
- PMCID: PMC12568640
- DOI: 10.3389/fmicb.2025.1579154
Virulence and mutations analysis based on the whole genome of a Brazilian Corynebacterium diphtheriae strain isolated from a cutaneous infection
Abstract
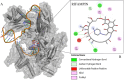
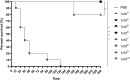
Corynebacterium diphtheriae is the main etiological agent of diphtheria, a potentially fatal disease whose most severe signs and symptoms result from the action of an exotoxin, the diphtheria toxin (DT). Although non-toxigenic C. diphtheriae strains have been associated with several diseases, including cutaneous infections and endocarditis, they are not monitored in many countries, and their mechanisms of virulence and antimicrobial resistance remain underexplored. Therefore, this study aimed to provide a comprehensive characterization -through genomic, in vitro, and in vivo analyses - of a non-toxigenic C. diphtheriae strain (46855) isolated from a leg lesion, highlighting its pathogenic potential and resistance profile. The isolate was assigned to a novel sequence type (ST-925) and was found to be resistant to tetracycline and rifampin. Multiple antimicrobial resistance genes were predicted in the genome, such as tet(33), rbpA, and rpoB2, in addition to mutations in the rpoB gene. A diverse set of virulence-associated genes related to adhesion, iron uptake systems, gene regulation, and post-translational modification was also identified. The isolate was able to form biofilm in vitro and exhibited strong virulence in Galleria mellonella larvae and A549 human pneumocyte cells. Finally, the structural analysis of the rpoB gene, carried out for the first time in this study, linked the observed mutations to rifampin resistance in C. diphtheriae. In summary, the data revealed that C. diphtheriae 46855, although non-toxigenic, harbors multiple genes associated with antimicrobial resistance and virulence, emphasizing the need for greater surveillance and functional studies on non-toxigenic strains.
Keywords: CRISPR-Cas system; Corynebacterium diphtheriae complex; non-toxigenic; resistance genes; virulence factors.
Copyright © 2025 Araújo, Santos, Prates, Perini, Silva, Ramos, Bokermann, Sacchi, Mattos Guaraldi, Campos, Cardoso, Castro, Silva, Sousa, Vieira, Santos, Camargo, Andrade, Silva, Sant’Anna, Viana and Azevedo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures






References
-
- Abraham M., Murtola T., Schulz R., Páll S., Smith J., Hess B., et al. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Africa Centres for Disease Control and Prevention. (2023). Diphtheria outbreak in Africa: Strengthening response capacities. Available online at: https://africacdc.org/news-item/diphtheria-outbreak-in-africa-strengthen... (accessed October 11, 2023).
LinkOut - more resources
Full Text Sources

