Artificial intelligence-powered spatial analysis of tumor microenvironment in patients with non-small cell lung cancer with acquired resistance to EGFR tyrosine kinase inhibitor
- PMID: 41173494
- PMCID: PMC12581073
- DOI: 10.1136/jitc-2025-012374
Artificial intelligence-powered spatial analysis of tumor microenvironment in patients with non-small cell lung cancer with acquired resistance to EGFR tyrosine kinase inhibitor
Abstract
Purpose: This study evaluated the dynamic changes in the tumor microenvironment (TME) in patients with non-small cell lung cancer (NSCLC) and acquired resistance to epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) using an artificial intelligence (AI)-powered spatial TME analyzer. We then assessed the predictive efficacy of immune-checkpoint inhibitors (ICIs)-based treatment.
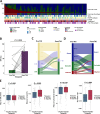
Experimental design: An AI-powered whole-slide image analyzer was used to segment cancer areas (CAs) and cancer stroma and to identify tumor-infiltrating lymphocytes (TILs), tertiary lymphoid structures, fibroblasts, and endothelial cells (ECs) in the tumor tissue. We analyzed 143 NSCLC samples after resistance to EGFR-TKIs from two cohorts: (1) 89 patients treated with ICI monotherapy and (2) 54 patients from the ATTLAS phase III trial comparing atezolizumab plus bevacizumab, paclitaxel, and carboplatin (ABCP) versus pemetrexed plus carboplatin.
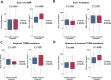
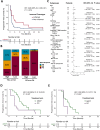
Results: Post-TKI samples showed reduced TILs in the CA (p=0.045) and increased ECs in the CA (p=0.005) compared with pre-TKI samples. These changes differed according to EGFR mutation subtype. Higher TILs in CA were associated with a better overall response rate (ORR) and progression-free survival (PFS). Similarly, higher EC levels in CA correlated with improved ORR and PFS. In the ATTLAS cohort, these factors were associated with clinical benefits from ABCP, with a significant association with TILs and a marginal association with ECs.
Conclusion: Our findings suggest that EGFR-TKIs affect the immune landscape of patients with EGFR-mutated NSCLC. Higher TILs or ECs in the CA were significantly associated with a favorable response to subsequent ICI-based treatment.
Trial registration number: NCT03991403.
Keywords: biomarker; lung cancer; tumor microenvironment - TME.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: Nothing directly related to this work. Outside of this work, SH reports employment at Lunit. HAJ reported advisory roles with Yuhan, Guardant, and AIMEDBIO, and received research funding from Yuhan. JSA received honoraria from Pfizer, Roche, BC World Pharmaceutical, Yuhan, Hanmi, Novartis, JW Pharmaceutical, Amgen, Boehringer Ingelheim, Menarini, Kyowa Kirin, AstraZeneca, Bayer, Lilly, Takeda, Boryung, and Samyang; and held advisory roles with Bayer, Yooyoung Pharmaceutical, Pharmbio Korea, Guardant Health, Yuhan, ImmuneOncia, Therapex, Daiichi Sankyo Korea, and Roche. M-JA received honoraria from AstraZeneca, Lilly, MSD, Takeda, Amgen, Merck Serono, and Yuhan; held advisory roles with AstraZeneca, Lilly, MSD, Takeda, Alpha Pharmaceutical, Amgen, Merck Serono, Pfizer, Yuhan, and Arcus Ventures; and received research funding from Yuhan. S-HL received honoraria from AstraZeneca/MedImmune, Roche, Merck Sharp & Dohme, Eli Lilly, Amgen, Abion, Daiichi Sankyo, and Yuhan; held consulting or advisory roles with AstraZeneca, Roche, Merck Sharp & Dohme, Pfizer, Eli Lilly, BMS/Ono, Daiichi Sankyo, Takeda, Janssen, IMBdx, Abion, Beigene, and ImmuneOncia; received research funding from Merck Sharp & Dohme, AstraZeneca, and Lunit; and received travel, accommodations, and expenses from Novartis. JWO, SH, CHA, SA, and C-YO are employees of Lunit.
Figures


References
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
