Evaluation of (Z)-endoxifen as a potential therapy for glioblastoma multiforme through computational and experimental analyses
- PMID: 41173946
- PMCID: PMC12578925
- DOI: 10.1038/s41598-025-22034-x
Evaluation of (Z)-endoxifen as a potential therapy for glioblastoma multiforme through computational and experimental analyses
Abstract
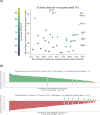
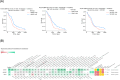
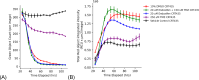
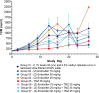
(Z)-endoxifen (endoxifen) is the active metabolite of tamoxifen. Endoxifen is a potent antiestrogen that binds and blocks estrogen receptor alpha (ERα) and estrogen receptor beta (ERβ). Early-phase clinical trials have shown that endoxifen has promising effects in patients with hormone-resistant metastatic breast cancer and other estrogen receptor-positive (ERα+) tumors. In addition, endoxifen has known estrogen-independent effects, such as inhibiting protein kinase C beta (PKCβ1). Given its broader mechanisms and demonstrated clinical activity with potential advantages over tamoxifen in breast cancer, endoxifen warrants investigation in other cancer types. This study aimed to identify new oncology indications with high therapeutic potential for endoxifen, as monotherapy or in combination, by applying the AI-powered PandaOmics platform to analyze a wide range of cancer types based on its mechanisms of action (MOA). Glioblastoma multiforme (GBM) emerged as a top candidate for endoxifen's therapeutic potential. In vitro studies in the CRT435 GBM cell line confirmed that endoxifen treatment reduced cell proliferation and induced cell death, while in vivo studies in a subcutaneous CRT435 patient-derived xenograft (PDX) model demonstrated a tolerable safety profile but no significant tumor growth reduction, likely reflecting limitations of the model used. This study underscores the application of AI-driven computational approaches in identifying new therapeutic hypotheses and demonstrates the potential of repurposing endoxifen for GBM treatment.
Keywords: (Z)-endoxifen; Artificial intelligence; Glioblastoma multiforme; Mechanism of action.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Insilico Medicine is a company developing an AI-based end-to-end integrated pipeline for drug discovery and development and engaged in aging and cancer research. AS, AV, KMA, AU, MK, AZ are affiliated with Insilico Medicine. SQ, HLR, and SSH each hold shares in Atossa Therapeutics, Inc. but declare no non-financial competing interests. HLR is lead independent director of Atossa Therapeutics, Inc.
Figures






References
-
- Zhavoronkov, A. et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol.37, 1038–1040 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

