Perioperative pembrolizumab in early-stage non-small cell lung cancer (NSCLC): conventional and distribution-based immune profiling of the tumor microenvironment and peripheral circulation
- PMID: 41176317
- PMCID: PMC12581070
- DOI: 10.1136/jitc-2025-012432
Perioperative pembrolizumab in early-stage non-small cell lung cancer (NSCLC): conventional and distribution-based immune profiling of the tumor microenvironment and peripheral circulation
Abstract
Purpose: A recently published phase 2 neoadjuvant trial in patients with early-stage non-small cell lung cancer (NSCLC) (NCT02818920) evaluated the potential efficacy of pembrolizumab administration in the absence of chemotherapy. This communication reports on conventional and distribution-based immune profiling efforts in efforts to identify novel biomarkers predictive of benefit.
Methods: Patients with stage 1B-3A NSCLC received two cycles of pembrolizumab (P), followed by surgical resection of the remaining tumors (NCT02818920). Banked peripheral blood mononuclear cells (PBMCs) were analyzed at baseline and following the second dose of P. Resected tumors were disaggregated, and cells were viably cryopreserved. Based on pathologic examination of the tumors, patients were categorized as major pathologic responders (MPR; ≤10% viable tumor present), or non-MPR (>10% viable tumor present). High-parameter immune phenotyping by flow cytometry was performed on all available tumor and PBMC specimens, and results were expressed using both conventional phenotypic frequency analyses as well as a novel distribution-based biomarker identification strategy aimed at discovery of patterns associated with MPR.
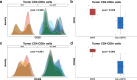
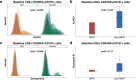
Results: Conventional, frequency-based flow cytometric immune phenotyping of participant tumor microenvironments and PBMC revealed several MPR-associated trends, only a few of which reached statistical significance. The distribution-based biomarker identification strategy greatly enhanced the discovery of statistically significant cell types and patterns of change associated with MPR.
Conclusions: This novel, distribution-based analytic framework identified MPR-associated immune cell subsets in baseline PBMC that were not evident using conventional frequency-based immune profiling. Profiling the microenvironment of MPR-associated tumors revealed statistically significant distributional differences among highly expressed cellular markers on CD8+ cells.
Keywords: Biomarker; Immunotherapy; Lung Cancer; Major pathologic response - MPR.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
