Transcriptomic Diversity of Pediatric Acute Myeloid Leukemia Genetic Drivers Correlates With Clinical Outcome and Expression of Stemness-Related Genes
- PMID: 41178390
- PMCID: PMC12580620
- DOI: 10.1002/cam4.71325
Transcriptomic Diversity of Pediatric Acute Myeloid Leukemia Genetic Drivers Correlates With Clinical Outcome and Expression of Stemness-Related Genes
Abstract
Background: Pediatric acute myeloid leukemia (pAML) is comprised of a diverse set of oncogenic drivers (ODs) that have been risk-stratified to inform prognosis and therapeutic decision-making. Despite proteomic, transcriptomic, genetic, and epigenetic characterization of the pAML landscape, questions still remain about why certain ODs have poorer prognoses than others.
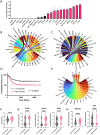
Methods: We analyze a large pAML bulk-RNA dataset (n = 435) and organize ODs along an axis of transcriptomic diversity by calculating the Simpson Diversity Index (SDI) of individual ODs.
Results: When comparing patients with low diversity ODs to patients with high diversity ODs, we observe poorer overall survival (HR = 1.877, 95% CI: 1.377-2.558, p = 0.0002) among patients harboring high diversity ODs in addition to an enrichment of stemness-related genes. We observe poorer survival of patients with high diversity ODs even when comparing patients with similar transcriptomic profiles (HR = 3.443, 95% CI: 1.817-6.525, p = 0.0028).
Conclusion: We identify a link between transcriptomic diversity, expression of stemness-related genes, and clinical outcome. Higher transcriptomic heterogeneity exhibited by high diversity ODs warrants further attention when identifying patients who can benefit from novel or high-intensity therapy.
Keywords: pediatric AML; transcriptomic diversity; transcriptomics.
© 2025 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
Q.R.B., E.S., R.M.R., C.L.M., and A.C. are inventors on a recently filed patent application related to separate CAR T‐related work. Additionally, A.C. discloses financial interests in the following entities working on antibody‐based conditioning approaches: Beam Therapeutics, Editas Medicines, GV, Inograft Biotherapeutics, Kyowa Kirin, and Prime Medicines. In addition, she is an inventor on antibody‐based conditioning patents licensed to Jasper Therapeutics, Gilead Sciences, Inograft Biotherapeutics, and Magenta Therapeutics. C.L.M. and E.S. are coinventors on multiple patents related to CAR T. C.L.M. is a cofounder, consults, and holds equity in CARGO Therapeutics, Link Cell Therapies, and GBM NewCo. C.L.M. consults for Ensoma and Immatics and received research funding from Tune and Lyell Immunopharma. E.S. consults for and holds equity in Lyell Immunopharma, and consults for Lepton Pharmaceuticals and Galaria.
Figures

References
-
- Ng S. W. K., Mitchell A., Kennedy J. A., et al., “A 17‐Gene Stemness Score for Rapid Determination of Risk in Acute Leukaemia,” Nature 540, no. 7633 (2016): 433–437. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

