Stereochemistry-aware string-based molecular generation
- PMID: 41190212
- PMCID: PMC12582147
- DOI: 10.1093/pnasnexus/pgaf329
Stereochemistry-aware string-based molecular generation
Abstract
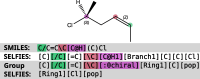
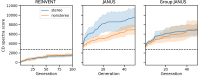
This study investigates the impact of incorporating stereochemical information, a crucial aspect of computational drug discovery and materials design, in molecular generative modeling. We present a detailed comparison of stereochemistry-aware and conventionally stereochemistry-unaware string-based generative approaches, utilizing both genetic algorithms and reinforcement learning-based techniques. To evaluate these models, we introduce novel benchmarks specifically designed to assess the importance of stereochemistry-aware generative modeling. Our results demonstrate that stereochemistry-aware models generally perform on par with or surpass conventional algorithms across various stereochemistry-sensitive tasks. However, we also observe that in scenarios where stereochemistry plays a less critical role, stereochemistry-aware models may face challenges due to the increased complexity of the chemical space they must navigate. This work provides insights into the trade-offs involved in incorporating stereochemical information in molecular generative models and offers guidance for selecting appropriate approaches based on specific application requirements.
Keywords: drug design; generative modeling; machine learning; molecular generation; stereochemistry.
© The Author(s) 2025. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures
References
-
- Sanchez-Lengeling B, Aspuru-Guzik A. 2018. Inverse molecular design using machine learning: generative models for matter engineering. Science. 361(6400):360–365. - PubMed
-
- De Cao N, Kipf T. 2018. Molgan: an implicit generative model for small molecular graphs [preprint], arXiv, arXiv:1805.11973. 10.48550/arXiv.1805.11973 - DOI
-
- Meyers J, Fabian B, Brown N. 2021. De novo molecular design and generative models. Drug Discov Today. 26(11):2707–2715. - PubMed
LinkOut - more resources
Full Text Sources

