EyaHOST, a modular genetic system for investigation of intercellular and tumor-host interactions in Drosophila melanogaster
- PMID: 41192417
- PMCID: PMC12664959
- DOI: 10.1016/j.crmeth.2025.101220
EyaHOST, a modular genetic system for investigation of intercellular and tumor-host interactions in Drosophila melanogaster
Abstract
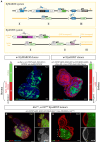
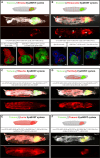
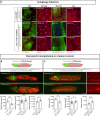
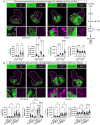
Studying intercellular and interorgan interactions in animal models is key to understanding development, physiology, and disease. We introduce EyaHOST, a system for clonal combinatorial loss- and gain-of-function genetics in fluorescently labeled cells under QF2-QUAS eya promoter control. Distinct from mosaic analysis with a repressible cell marker (MARCM), it reserves the use of genome-wide GAL4-UAS tools to manipulate any host tissue. EyaHOST-driven RasV12 overexpression with scribble knockdown recapitulates key cancer features, including systemic catabolic switching and organ wasting. We demonstrate effective tissue-specific manipulation of host compartments, including homotypic epithelial neighbors, immune cells, fat body, and muscle. Organ-specific inhibition of autophagy or stimulation of growth signaling via PTEN knockdown in fat body or muscle prevents cachexia-like wasting. Additionally, tumors trigger caspase-driven apoptosis in the neighboring epithelium, and blocking apoptosis with p35 enhances tumor growth. EyaHOST provides a modular platform to dissect mechanisms of intercellular and interorgan communication under physiological or disease conditions.
Keywords: CP: cancer biology; CP: genetics; Drosophila; QF2; Ras; Scrib; apoptosis-induced proliferation; cachexia; cancer model; cell competition; tumor-host; tumorigenesis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Update of
-
EyaHOST, a modular genetic system for investigation of intercellular and tumor-host interactions in Drosophila melanogaster.bioRxiv [Preprint]. 2024 Sep 11:2024.09.06.611647. doi: 10.1101/2024.09.06.611647. bioRxiv. 2024. Update in: Cell Rep Methods. 2025 Nov 17;5(11):101220. doi: 10.1016/j.crmeth.2025.101220. PMID: 39314415 Free PMC article. Updated. Preprint.
References
-
- Golic K.G., Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

