Structural and functional characterization of TgGSK3, a druggable kinase in Toxoplasma gondii
- PMID: 41193440
- PMCID: PMC12589562
- DOI: 10.1038/s41467-025-64701-7
Structural and functional characterization of TgGSK3, a druggable kinase in Toxoplasma gondii
Abstract
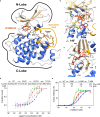
Toxoplasma gondii and Cryptosporidium species are apicomplexan parasites of significant medical and veterinary importance. Although current therapeutic options for toxoplasmosis and cryptosporidiosis demonstrate notable efficacy, their clinical efficacy is often limited by suboptimal efficacy and frequent adverse effects. Moreover, therapeutic alternatives remain limited or nonexistent, particularly for cryptosporidiosis, for which nitazoxanide is currently the only approved medication to treat diarrhea in adults and children older than 1 year of age. To identify alternative therapeutic options for addressing these health challenges, we performed a phenotypic screening of an FDA-approved drug repurposing library against Toxoplasma. This screening identifies LY2090314 as a potent inhibitor of T. gondii and Cryptosporidium growth in mammalian cells. Through a target deconvolution strategy combining forward genetics, transcriptome sequencing, and computational mutation analysis, we elucidate the parasiticidal mechanism of LY2090314 and demonstrate that TgGSK3 kinase is its primary molecular target. We also report the first X-ray crystal structure of LY2090314 bound to TgGSK3, resolved at 2.1 Å, which reveals an interaction mode characteristic of type I ATP-competitive inhibitors. Furthermore, interactome analysis uncovers functional connections between TgGSK3 and key cytoskeletal and signaling regulators, providing insights into compound's effects. Collectively, these findings validate TgGSK3 as a promising therapeutic target for toxoplasmosis and offer mechanistic insights into apicomplexan GSK3 biology.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures




References
-
- World malaria report. https://www.who.int/teams/global-malaria-programme/reports/world-malaria... (2023).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

