CDK7-targeted therapy effectively disrupts cell cycle progression and oncogenic signaling in head and neck cancer
- PMID: 41193441
- PMCID: PMC12589458
- DOI: 10.1038/s41392-025-02452-z
CDK7-targeted therapy effectively disrupts cell cycle progression and oncogenic signaling in head and neck cancer
Abstract
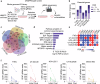
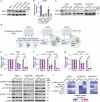
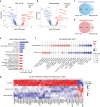
Head and neck squamous cell carcinoma (HNSCC) remains a prevalent and aggressive malignancy, characterized by a lack of targeted therapies and limited clinical benefits. Here, we conducted an optimized whole-genome CRISPR screen across five HNSCC cell lines aimed at identifying actionable genetic vulnerabilities for rapid preclinical evaluation as novel targeted therapies. Given their critical role in cancer, cyclin-dependent kinases (CDKs) were prioritized for further investigation. Among these, CDK7 was identified as an essential and targetable gene across all five cell lines, prompting its selection for in-depth functional and molecular characterization. Genetic and pharmacological inhibition of CDK7 significantly and consistently reduced tumor cell proliferation due to generalized cell cycle arrest and apoptosis induction. Additionally, CDK7 knockout (KO) and selective inhibitors (YKL-5-124 and samuraciclib) demonstrated potent antitumor activity, effectively suppressing tumor growth in HNSCC patient-derived organoids (PDOs), as well as in both cell line- and patient-derived xenograft (PDX) mouse models with minimal toxicity. Mechanistically, CDK7 inhibition led to a broad downregulation of gene sets related to cell cycle progression and DNA repair, and significantly reduced the transcription of essential genes and untargetable vulnerabilities identified by our CRISPR screen. These findings highlight CDK7 as a promising therapeutic target for HNSCC. Our study provides strong evidence of the robust antitumor activity of CDK7-selective inhibition in disease-relevant preclinical models, strongly supporting its progression to clinical testing.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.T. is a shareholder of and has received research funding from Storm Therapeutics Ltd. All the other authors declare that they have no competing interests.
Figures




References
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Burtness, B. et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet394, 1915–1928 (2019). - PubMed
-
- Bonner, J. A. et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J. Med.354, 567–578 (2006). - PubMed
MeSH terms
Substances
Grants and funding
- FI23/00037/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- CP23/00101/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- BP21-205/Gobierno del Principado de Asturias (Government of the Principality of Asturias)
- PI24/00398/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- WT_/Wellcome Trust/United Kingdom
- PI22/00167/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- PI19/00560/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- CB16/12/00228/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- CP24/00064/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- CB16/12/00390/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- IDI/2021/000079/Gobierno del Principado de Asturias (Government of the Principality of Asturias)
- PI21/00208/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- FI20/00064/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

