DHCR7 as a Prognostic and Immunological Biomarker in Human Pan-Cancer: A Comprehensive Evaluation
- PMID: 41198144
- PMCID: PMC12591708
- DOI: 10.1002/cnr2.70376
DHCR7 as a Prognostic and Immunological Biomarker in Human Pan-Cancer: A Comprehensive Evaluation
Abstract
Background: The 7-Dehydrocholesterol reductase (DHCR7), a critical enzyme catalyzing the final step of the cholesterol biosynthesis pathway, has gained attention for its potential role in tumorigenesis. This study systematically investigated the association between DHCR7 expression and oncogenic processes across multiple cancer types.
Methods: Multi-omics data were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) repositories. DHCR7 expression patterns were analyzed using Oncomine, TIMER, and GEPIA platforms. Prognostic significance was assessed via Kaplan-Meier plotter and GEPIA. Tumor stage correlations and immune/molecular subtype associations were evaluated using TISIDB. SangerBox facilitated analysis of DHCR7's associations with immune checkpoint (ICP) molecules, tumor mutational burden (TMB), microsatellite instability (MSI), mutant-allele tumor heterogeneity (MATH), neoantigen load, and immune cell infiltration.
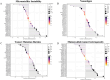
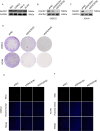
Results: DHCR7 exhibited significant overexpression in most malignancies, correlating with advanced tumor stage (p < 0.05), metastatic progression, and reduced overall survival (HR = 1.34, 95% CI: 1.18-1.52). Strong associations emerged between DHCR7 expression and critical immunomodulatory parameters: positive correlations with ICPs (PD-L1: r = 0.62, CTLA4: r = 0.58). Significant links to TMB (p = 2.1e-5), MSI (p = 4.3e-4), and MATH (p = 7.8e-3). Distinct immune infiltration patterns, particularly in bladder carcinoma (BLCA), renal clear cell carcinoma (KIRC), and prostate adenocarcinoma (PRAD). Co-expression network analysis revealed DHCR7's involvement in immune response regulation (GO:0002764, FDR = 0.003), leukocyte differentiation (GO:0002521, FDR = 0.012), and angiogenesis (GO:0001525, FDR = 0.018).
Conclusions: These pan-cancer analyses identify DHCR7 as a multifaceted biomarker with dual prognostic and immunotherapeutic relevance. Its involvement in tumor immune microenvironment modulation suggests potential as a therapeutic target.
Keywords: DHCR7; Pan‐cancer analysis; immunotherapy; prognostic biomarker; tumor microenvironment.
© 2025 The Author(s). Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures











References
-
- Kim J. H., Lee J. N., and Paik Y. K., “Cholesterol Biosynthesis From Lanosterol. A Concerted Role for Sp1 and NF‐Y‐Binding Sites for Sterol‐Mediated Regulation of Rat 7‐Dehydrocholesterol Reductase Gene Expression,” Journal of Biological Chemistry 276, no. 21 (2001): 18153–18160. - PubMed
-
- Kritzer A., Dutta R., Pramparo T., Terner‐Rosenthal J., Vig P., and Steiner R. D., “Smith‐Lemli‐Opitz Syndrome: Clinical, Biochemical, and Genetic Insights With Emerging Treatment Opportunities,” Genetics in Medicine 27 (2025): 101450. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

