Targeting polyamine metabolism and ferroptosis enhances the efficacy of KRAS-targeted therapy depending on KEAP1 status
- PMID: 41219185
- PMCID: PMC12606100
- DOI: 10.1038/s41467-025-65441-4
Targeting polyamine metabolism and ferroptosis enhances the efficacy of KRAS-targeted therapy depending on KEAP1 status
Abstract
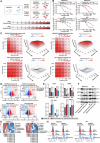
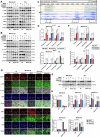
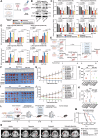
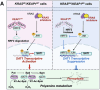
The resistance to KRAS-targeted therapies, particularly due to co-occurring gene mutations, remains a significant challenge. Through a metabolite library screening, we reveal that polyamines sensitize KRAS inhibitors only in KRASMU/KEAP1WT cells but not in KRASMU/KEAP1MU cells. Transcriptome sequencing and metabolome profiling pinpoint SAT1, the key enzyme in polyamine metabolism, as essential for this divergence. In KRASMU/KEAP1WT context, treatment of KRAS inhibitors activates JNK/c-Jun pathway and SAT1 expression, while the augmented SAT1 facilitates polyamine metabolism and KRAS inhibitors-induced ferroptosis. Conversely, in KRASMU/KEAP1MU cells, activated JNK promotes the degradation of NRF2, thereby inhibiting SAT1 expression. Our results further demonstrate that polyamine supplementation enhances KRAS-targeted therapy in KRASMU/KEAP1WT resistant cells, patient-derived organoids, xenografts, and spontaneously tumorigenic mice, while KRASMU/KEAP1MU models require lentivirus or adeno-associated virus-mediated SAT1 overexpression prior to polyamine treatment, to augment ferroptosis and drug sensitivity. Our findings highlight SAT1-mediated polyamine metabolism as a promising target in precision treatments for KRAS-mutant cancers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Li, S., Balmain, A. & Counter, C. M. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat. Rev. Cancer18, 767–777 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

