Persistent mirusvirus infection in the marine protist Aurantiochytrium
- PMID: 41219190
- PMCID: PMC12606086
- DOI: 10.1038/s41467-025-65172-6
Persistent mirusvirus infection in the marine protist Aurantiochytrium
Abstract
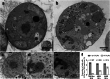
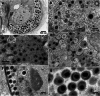
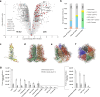
Mirusviruses are abundant and broadly distributed double-stranded (ds) DNA viruses recently discovered in marine metagenomic data. Their host range and ecological impact are unclear. The protist Aurantiochytrium limacinum possesses two mirusvirus-like genomic elements, one a circular episome (AurliV-1) and the other (AurliV-2) a chromosomal integrant. Here we show that genes in both genomes are expressed and viral particles containing mainly AurliV-1 DNA are produced under starvation conditions and when cells are cultured in standard growth medium. We detected viral particles of ~140 nm in the nucleus, in cytoplasmic vesicles, between the plasma membrane and cell wall, and in the extracellular environment. Of 67 AurliV-1-encoded proteins detected using proteomics, 45 are enriched under starvation conditions, including the structurally important major capsid and triplex proteins. Our results establish Aurantiochytrium as a model system for elucidating mirusvirus-host interactions and demonstrate persistent viral infection in a microbial eukaryote.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

