Bioinformatics and experimental animal model reveal the prognostic value of immunogenic cell death-related proteins in idiopathic pulmonary fibrosis
- PMID: 41229825
- PMCID: PMC12603469
- DOI: 10.21037/jtd-2025-616
Bioinformatics and experimental animal model reveal the prognostic value of immunogenic cell death-related proteins in idiopathic pulmonary fibrosis
Abstract
Background: Immunogenic cell death (ICD) is a type of regulated cell death (RCD) that activates adaptive immune responses and shapes the immune microenvironment. Its role in idiopathic pulmonary fibrosis (IPF), a progressive and fatal lung disease, remains unclear. This study aims to identify ICD-related gene signatures and evaluate their prognostic value in IPF through bioinformatic analysis and experimental validation.
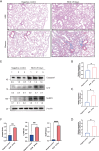
Methods: Gene expression profiles and clinical data from 176 IPF patients and 20 healthy controls were obtained from the GSE70866 dataset. A set of 34 ICD-related genes was curated from literature. Differential expression analysis, univariate Cox regression, and least absolute shrinkage and selection operator (LASSO)-penalized Cox regression were used to identify prognostic genes and construct a risk model. The model was validated internally and using an independent cohort (GSE70867). Immune cell infiltration was assessed via CIBERSORT. Expression of identified genes was further validated in a bleomycin-induced pulmonary fibrosis mouse model using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) and Western blot.
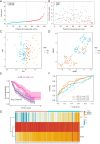
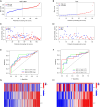
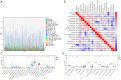
Results: Ten ICD-related genes were differentially expressed in IPF patients and associated with prognosis. A three-gene prognostic signature (IL10, CASP1, NLRP3) was established. Patients were stratified into high- and low-risk groups with significantly different overall survival (P<0.05). The risk score proved to be an independent prognostic factor for IPF. Time-dependent receiver operating characteristic (ROC) analysis showed strong predictive performance for 1-, 2-, and 3-year survival. Immune profiling revealed significant differences in mast cells, natural killer cells, and dendritic cells between risk groups. In the mouse model, mRNA and protein expression of IL10, CASP1, and NLRP3 were significantly upregulated in fibrotic lungs.
Conclusions: We developed and validated a novel ICD-related gene signature capable of predicting prognosis in IPF patients. The three-gene risk model may serve as a promising tool for risk stratification and personalized treatment planning in IPF.
Keywords: Immunogenic cell death (ICD); biomarkers; idiopathic pulmonary fibrosis (IPF); prognostic model.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-2025-616/coif). G.Z. reports that this study was supported by the Key Clinical Research Projects of The First Affiliated Hospital of Xi’an Jiaotong University (No. XJTU1AF-CRF-2023-006) and Xi’an Jiaotong University Basic-Clinical Integration Innovation Program (No. YXJLRH2022033). Y.Z. reports that this study was supported by Institutional Foundation of The First Affiliated Hospital of Xi’an Jiaotong University (No. 2024-MS-15). The other authors have no conflicts of interest to declare.
Figures