Unveiling the trophic dynamics and ecological roles of demersal fish in Hong Kong: A metabarcoding and isotope analysis approach
- PMID: 41231845
- PMCID: PMC12614624
- DOI: 10.1371/journal.pone.0335343
Unveiling the trophic dynamics and ecological roles of demersal fish in Hong Kong: A metabarcoding and isotope analysis approach
Abstract
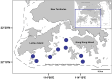
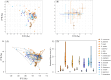
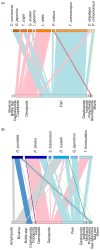
This study provides a comprehensive examination of the trophic ecology and feeding dynamics of 16 demersal fish species inhabiting the southern and southwestern waters of Hong Kong, utilizing advanced 12S and COI gut content metabarcoding alongside stable isotope analysis. Dietary dissimilarities, primarily driven by Decapoda and fish, are significant among species. Network plots further highlight unique predator-prey interactions. Eight species, including horn dragonet Callionymus curvicornis and Japanese butterflyray Gymnura japonica, are identified as piscivores, primarily preying on demersal fish, while six species such as rough flathead Grammoplites scaber and Japanese flathead Inegocia japonica are classified as crustacean feeders, focusing on Decapoda. Notably, spotted sicklefish Drepane punctata and goatee croaker Dendrophysa russelii exhibit unique feeding behaviors, relying on brittle star and Bivalvia, respectively, and demonstrate non-selective feeding patterns that do not prioritize dominant environmental species. This diverse range of prey consumption highlights the critical roles these fish play in regulating demersal fish and benthic invertebrate communities. The study also reveals clear trophic niche partitioning with low isotope niche overlap, predominantly below 55.30%, except for a notable overlap of 72.91% between bartail flathead Platycephalus indicus and goatee croaker D. russelii. Our results established essential baseline data on trophic niche diversification and resource partitioning through varied dietary preferences, facilitating coexistence and resilience within the ecosystem. This research serves as a foundational assessment of the trophic dynamics and ecological stability in Hong Kong's marine ecosystems, offering valuable insights into anthropogenic pressures and guiding the development of specific conservation strategies aimed at preserving fish biodiversity and sustaining global fisheries.
Copyright: © 2025 Wong et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Macarthur R, Levins R. The Limiting Similarity, Convergence, and Divergence of Coexisting Species. The American Naturalist. 1967;101(921):377–85. doi: 10.1086/282505 - DOI
-
- Tilman D. The Importance of the Mechanisms of Interspecific Competition. The American Naturalist. 1987;129(5):769–74. doi: 10.1086/284672 - DOI
-
- Ross ST. Resource Partitioning in Fish Assemblages: A Review of Field Studies. Copeia. 1986;1986(2):352. doi: 10.2307/1444996 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

