Microbial signals in primary and metastatic brain tumors
- PMID: 41238915
- PMCID: PMC12618227
- DOI: 10.1038/s41591-025-03957-4
Microbial signals in primary and metastatic brain tumors
Abstract
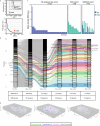
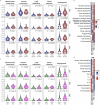
Gliomas and brain metastases are associated with poor prognosis, necessitating a deeper understanding of brain tumor biology and the development of effective therapeutic strategies. Although our group and others have demonstrated microbial presence in various tumors, recent controversies regarding cancer-type-specific intratumoral microbiota emphasize the importance of rigorous, orthogonal validation. This prospective, multi-institutional study included a total of 243 samples from 221 patients, comprising 168 glioma and brain metastases samples and 75 non-cancerous or tumor-adjacent tissues. Using stringent fluorescence in situ hybridization, immunohistochemistry and high-resolution spatial imaging, we detected intracellular bacterial 16S rRNA and lipopolysaccharides in both glioma and brain metastases samples, localized to tumor, immune and stromal cells. Custom 16S and metagenomic sequencing workflows identified taxa associated with intratumoral bacterial signals in the tumor microenvironment; however, standard culture methods did not yield readily cultivable microbiota. Spatial analyses revealed significant correlations between bacterial 16S signals and antimicrobial and immunometabolic signatures at regional, neighborhood and cellular levels. Furthermore, intratumoral 16S bacterial signals showed sequence overlap with matched oral and gut microbiota, suggesting a possible connection with distant communities. Together, these findings introduce microbial elements as a component of the brain tumor microenvironment and lay the foundation for future mechanistic and translational studies.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: R.R.J. serves as a consultant and provides expert testimony for MaaT Pharma and Postbiotics Plus and receives patent license fees from Seres Therapeutics and stock options from Seres Therapeutics and Postbiotics Plus. Unrelated to this work, P.K.B. has received consultant fees from Atavistik, Advise Connect Inspire, AngioChem, Axiom Healthcare Strategies, CraniUS, Dantari, ElevateBio, Eli Lilly, Genentech, InCephalo Therapeutics, Kazia, Merck, MPM Capital, Pfizer, Sintetica, SK Life Science, Tesaro and Voyager Therapeutics; equity from Selectin Therapeutics; speaker honoraria from Eli Lilly, Genentech, Medscape and Merck; and institutional research funding (to Massachusetts General Hospital) from Eli Lilly, Kinnate Biopharma, Merck and Mirati. M.A.D. has been a consultant to Roche/Genentech, Array, Pfizer, Novartis, Bristol Myers Squibb, GlaxoSmithKline, Sanofi-Aventis, Vaccinex, Apexigen, Eisai, Iovance, Merck and ABM Therapeutics, and he has been the principal investigator of research grants to MD Anderson by Roche/Genentech, GlaxoSmithKline, Sanofi-Aventis, Merck, Myriad, Oncothyreon, Pfizer, ABM Therapeutics and LEAD Pharma. N.T. is founder and holder of equity in BrainDynamics, Inc. and also holds an equity position in Nervonik. V.K.P. served as an advisory board member and received honoraria from Orbus Therapeutics, Insightec, Novocure, Boehringer Ingelheim, Telix Pharma, Tango Pharmaceuticals, Bayer and Servier; received research support from Bexion, Karyopharm, Remedy Plan Therapeutics, Radiomedix, Merck and Servier; and holds equity in Gilead. J.A.W. reports compensation for speaker’s bureau and honoraria from PeerView and serves as a consultant and/or advisory board member for Gustave Roussy Cancer Center, OSE Immunotherapeutics, Bayer Therapeutics, James Cancer Center OSU and Daiichi Sankyo. The remaining authors declare no competing interests.
Figures










References
-
- Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov.12, 31–46 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

