Does polypharmacy affect epigenetic aging in older people? Evidence from a longitudinal epigenome-wide methylation study
- PMID: 41239444
- PMCID: PMC12619211
- DOI: 10.1186/s13148-025-02007-7
Does polypharmacy affect epigenetic aging in older people? Evidence from a longitudinal epigenome-wide methylation study
Abstract
Background: Polypharmacy, defined as taking ≥ 5 different daily medications, is common in older adults and has been linked with neuropsychiatric/neurological and other health conditions. To clarify the potential molecular implications, we tested the hypothesis that polypharmacy may influence DNA methylation (DNAm) patterns in aging, in a longitudinal Italian cohort (N = 1,098; mean (SD) age at recruitment: 58.8 (5.6) years, 51.3% women; median (IQR) follow-up 12.6 (1.1) years).
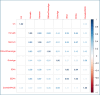
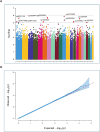
Results: We tested associations of polypharmacy with several DNAm aging clocks (Hannum, Horvath, GrimAge, DNAmPhenoAge, DunedinPACE), through linear mixed models incrementally adjusted for age, sex, education, prevalent health conditions and lifestyles, leukocyte counts and residual batch effects. This revealed significant positive associations of GrimAge acceleration and DunedinPACE with the switch to polypharmacy status during follow-up (Beta (SE): 0.024 (0.008) and0.0012 (0.0004)). While the association of GrimAge was driven by a DNAm-based surrogate of tissue inhibitor metalloproteinase 1 (TIMP-1), no significant association was detected for component CpGs of DunedinPACE. When we tested associations of polypharmacy with 668,413 CpGs epigenome-wide, we observed no statistically significant findings (top hit: cg07675998; chr11q13.1; Beta (SE) = 0.009 (0.002); p = 1.5 × 10-6). However, these showed significant enrichments of several biological functions and pathways related to renal tissue, lipoproteins, inflammatory and immune response.
Conclusions: These findings suggest an influence of polypharmacy on accelerated epigenetic aging and on altered methylation patterns in the genome, suggesting a potential implication of pathways related to renal tissue development, lipoproteins and cholesterol homeostasis, inflammatory and immune response, in line with previous proteomic analyses of polypharmacy mouse models. These observations also suggest potential targets for mitigating disruptive effects of polypharmacy on elderly health.
Keywords: Aging; Chronic health conditions; Elderly; Epigenetic clocks; Inflammation; Neurodegenerative and neuropsychiatric disease; Polypharmacy.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the IRCCS Neuromed (28/02/2018). All participants provided written informed consent. Competing interests: The authors declare no competing interests.
Figures
References
-
- A. Hung, Y. H. Kim, e J. M. Pavon, «Deprescribing in older adults with polypharmacy», BMJ, p. e074892, mag. 2024, 10.1136/bmj-2023-074892. - PubMed
-
- Nicholson K, et al. Prevalence of multimorbidity and polypharmacy among adults and older adults: a systematic review. Lancet Healthy Longev. 2024;5(4):e287–96. 10.1016/S2666-7568(24)00007-2. - PubMed
URLS
-
- Farmastat (source #1): https://www.farmastat.it/ (Accessed April 20, 2024)
-
- Farmastat (source #2): https://www.marnonet.it/mws_home.aspx (Accessed April 20, 2024)
-
- R software: https://www.r-project.org/ methylclock package: https://www.bioconductor.org/packages/release/bioc/html/methylclock.html
-
- PC-based clocks scripts: https://github.com/MorganLevineLab/PC-ClocksPC-based clocks scripts: https://github.com/MorganLevineLab/PC-Clocks
-
- DunedinPACE package: https://github.com/danbelsky/DunedinPACE/tree/main
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

