Bacterial natural transformation drives cassette shuffling and simplifies recombination in chromosomal integrons
- PMID: 41242524
- PMCID: PMC12614218
- DOI: 10.1093/nar/gkaf1162
Bacterial natural transformation drives cassette shuffling and simplifies recombination in chromosomal integrons
Abstract
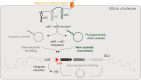
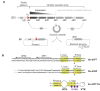
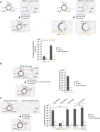
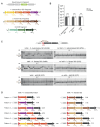
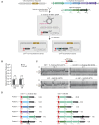
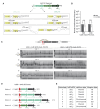
Integrons act as biobanks of gene cassettes conferring functions crucial for bacterial defense, including protection against phages and antibiotics. They enable bacterial on-demand adaptation through capture and shuffling of the cassettes under stress conditions. Our results underscore the significant role of horizontal gene transfer in integron cassette recombination. We discover that sedentary chromosomal integrons (SCIs), such as those found in Vibrio cholerae, efficiently excise and recruit cassettes from linear single-stranded DNA fragments acquired during natural transformation. We propose a simplified mechanism for the cassette excision process from this type of substrates, requiring only a single strand exchange at the attC recombination sites, ruling out any replicative mechanism. We also observe a higher specificity of the V. cholerae integrase for attC recombination sites from the V. cholerae repeat-type, a trait differentiating SCI integrases from the mobile integron (MI) ones. This specificity, likely stemming from a long-term co-evolution between SCI integrases and attC sites, impedes the recruitment of cassettes from phylogenetically distant integrons. Collectively, our findings may explain the greater attC site homogeneity observed in SCIs compared to MIs and showcase the role of natural transformation in driving cassette shuffling and simplifying the cassette recombination mechanism, thereby expanding bacterial phenotypic diversity.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
-
- Escudero JA, Loot C, Nivina A et al. The Integron: adaptation on demand. Microbiol Spectr. 2015;3:MDNA3–0019-2014. - PubMed
-
- Escudero JAL C, Mazel D. In: Rampelotto PH (ed.), Molecular Mechanisms of Microbial Evolution, 2018, 199–239.
MeSH terms
Substances
Grants and funding
- CNRS-UMR 3525/Institut Pasteur, the Centre National de la Recherche Scientifique
- EQU202103012569/Fondation pour la Recherche Médicale
- EQU201903007835/Fondation pour la Recherche Médicale
- FDT202404018553/Fondation pour la Recherche Médicale
- ANR-21-CE12-0002-01/Agence Nationale de la Recherche
- ANR-24-CE12-7883-01/Agence Nationale de la Recherche
- ANR-20-CE35-014/Agence Nationale de la Recherche
- ANR-10-LABX-62-IBEID/French Government's Investissement d'Avenir program Laboratoire d'Excellence 'Integrative Biology of Emerging Infectious Diseases'
- French Government
- PIA/ANR-16-CONV-0005 INCEPTION/Investissement d'Avenir program
- Direction Générale de l'Armement
LinkOut - more resources
Full Text Sources

