Automated quantification of Ki-67 expression in breast cancer from H&E-stained slides using a transformer-based regression model
- PMID: 41250202
- PMCID: PMC12625586
- DOI: 10.1186/s13058-025-02149-9
Automated quantification of Ki-67 expression in breast cancer from H&E-stained slides using a transformer-based regression model
Abstract
Background: Accurate quantification of the Ki-67 proliferation index is essential for breast cancer prognosis and treatment planning. Current automated methods, including classical and deep learning approaches based on cell detection or segmentation, often face challenges due to densely packed nuclei, morphological variability, and inter-laboratory differences. Since Hematoxylin and Eosin (H&E) staining is routinely performed, accurately estimating Ki-67 from these slides could save resources by eliminating the need for additional immunohistochemical (IHC) staining. We developed and validated a transformer-based regression model to estimate Ki-67 expression directly from H&E-stained Whole Slide Images (WSIs).
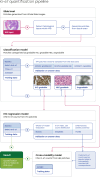
Methods: We used seven public datasets to select optimal transformer-based architectures and hyperparameters. WSIs underwent preprocessing to filter poor-quality patches, with a classification model identifying gradable patches. Only gradable patches proceeded to Ki-67 quantification. Initially, a regression model was trained on IHC-stained patches using independently annotated datasets, bypassing segmentation methods. This model generated pseudo-labels for unlabeled IHC patches, which were then paired with corresponding H&E images, with a separate model trained using only these H&E patches. Both models were evaluated separately across 1153 H&E and 843 IHC-stained WSIs, employing metrics such as R2.
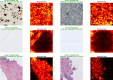
Results: Our regression model had good predictive accuracy, with R2 values exceeding 0.90 for quantifying positive cells, negative cells, and Ki-67 ratios. The classification model effectively distinguished gradable patches, achieving a near-perfect AUROC (~ 100%) across independent and unseen datasets. Cross-modality performance was robust, achieving R2 values over 0.95 for positive and negative cell counts. Additionally, the model accurately captured the proliferation patterns from H&E-stained WSIs.
Conclusion: Our approach precisely quantifies Ki-67 expression and automates hotspot detection from WSIs, providing a scalable tool for digital pathology workflows. The cross-modality model potentially quantifies molecular expression from morphological features using H&E-stained WSIs.
Keywords: Breast Cancer; Digital Pathology; Hematoxylin and Eosin; Immunohistochemistry; Ki-67 Index; Regression Analysis; Vision Transformer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: We incorporated seven publicly accessible histopathology datasets without direct patient interaction or personally identifiable information; no additional ethical approval was required. It complies with ethical guidelines, and all dataset sources follow data-sharing policies. Consent for publication: Not applicable. Competing interests: A.K.C., M.T.B., P.W.T., and A.W.H. are co-founders of Pandani Solutions Pty Ltd, Australia, specialising in computational pathology.
Figures




References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians [Internet]. 2024 May 1 [cited 2025 Feb 4];74(3):229–63. Available from: https://onlinelibrary.wiley.com/doi/abs/10.3322/caac.21834 - PubMed
-
- Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. The Breast : Official Journal of the European Society of Mastology [Internet]. 2022 Sep 2 [cited 2024 Nov 22];66:15. Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC9465273/ - PMC - PubMed
-
- Wojtyla C, Bertuccio P, Wojtyla A, La Vecchia C. European trends in breast cancer mortality, 1980–2017 and predictions to 2025. European journal of cancer (Oxford, England : 1990) [Internet]. 2021 Jul [cited 2025 Feb 4];152. Available from: https://pubmed.ncbi.nlm.nih.gov/34062485/ - PubMed
-
- Rudolph A, Chang-Claude J, Schmidt MK. Gene–environment interaction and risk of breast cancer. British Journal of Cancer [Internet]. 2016 Jan 12 [cited 2025 Apr 2];114(2):125–33. Available from: https://www.nature.com/articles/bjc2015439 - PMC - PubMed
-
- Obeagu EI, Obeagu GU. Breast cancer: A review of risk factors and diagnosis. Medicine [Internet]. 2024 Jan 19 [cited 2025 Apr 2];103(3). Available from: https://pubmed.ncbi.nlm.nih.gov/38241592/ - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

