This is a preprint.
Integrated Multiclass Driver ctDNA Profiling Enables MPNST Detection and Monitoring in NF1 Patients
- PMID: 41255978
- PMCID: PMC12622166
- DOI: 10.21203/rs.3.rs-7330245/v1
Integrated Multiclass Driver ctDNA Profiling Enables MPNST Detection and Monitoring in NF1 Patients
Abstract
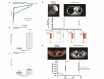
We developed and cross-validated an integrated circulating tumor DNA assay incorporating SNVs, indels, CNAs, and SVs to distinguish neurofibromatosis type 1 patients with malignant peripheral nerve sheath tumors from those with benign plexiform neurofibromas or tumor-free controls. Among 82 participants, the assay achieved an AUC of 0.904, compared with 0.735 for genome-wide CNAs. We also detected recurrent disease-specific driver alterations, relapse up to three months before diagnosis, and early clearance consistent with durable remission.
Conflict of interest statement
Declarations Competing Interests: P.A.J, J.J.S., R.T.S., J.F.S., A.A.C. and A.C.H. have patent filings related to cancer biomarkers. A.A.C. has licensed technology to Droplet Biosciences, Tempus Labs, and LiquidCell Dx. A.A.C. has served as a consultant/advisor to Roche, Tempus, Guardant Health, Exact Sciences, Caris, Geneoscopy, Illumina, Myriad Genetics, Invitae, Daiichi Sankyo, AstraZeneca, AlphaSights, DeciBio and Guidepoint. A.A.C. has received honoraria from Agilent and Illumina, and has received research support from Illumina, Roche and Tempus. A.A.C. has stock options in Geneoscopy, and ownership interests in Droplet Biosciences, LiquidCell Dx and CytoTrace Biosciences. A.C.H. has served on advisory boards for AstraZeneca/Alexion and Springworks Therapeutics. No potential conflicts of interest were disclosed by the other authors.
Figures

References
-
- Nguyen R., Dombi E., Widemann B. C., Solomon J., Fuensterer C., Kluwe L., Friedman J. M., & Mautner V. F. (2012). Growth dynamics of plexiform neurofibromas: a retrospective cohort study of 201 patients with neurofibromatosis 1. Orphanet journal of rare diseases, 7, 75. 10.1186/1750-1172-7-75 23035791 - DOI - PMC - PubMed
-
- Carton C., Evans D. G., Blanco I., Friedrich R. E., Ferner R. E., Farschtschi S., Salvador H., Azizi A. A., Mautner V., Röhl C., Peltonen S., Stivaros S., Legius E., Oostenbrink R., & ERN GENTURIS NF1 Tumour Management Guideline Group (2023). ERN GENTURIS tumour surveillance guidelines for individuals with neurofibromatosis type 1. EClinicalMedicine, 56, 101818. 10.1016/j.eclinm.2022.101818 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

