This is a preprint.
Pathogen-Phage Geomapping to Overcome Resistance
- PMID: 41256424
- PMCID: PMC12621808
- DOI: 10.1101/2025.10.01.679739
Pathogen-Phage Geomapping to Overcome Resistance
Abstract
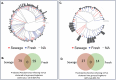
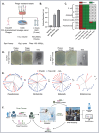
The rise of antibiotic resistance has renewed interest in bacteriophages as therapeutic alternatives. However, co-evolution of phage and bacteria will naturally give rise to phage-resistant pathogens, complicating phage therapy efforts. A critical bottleneck in the production of phage therapeutics is the discovery of virulent phages against resistant pathogens. Conventional methods for discovery are time-consuming, biased, and laborious, limiting the potential for identifying suitable phage candidates. To overcome these limitations, we combined small-volume environmental sampling with 16S rRNA sequencing to identify reservoirs where bacterial hosts co-exist with their phage predators. This strategy, which we term geographical phage mapping (geΦmapping), pinpoints ecological "hotspots" for targeted phage hunting. We further developed a portable phage hunting device (ΦHD) that generates highly enriched phage concentrates directly from these reservoirs. By integrating geΦmapping with high-throughput enrichment, we constructed the RΦ library, a diverse collection of novel phages targeting resistant pathogens.
Conflict of interest statement
Disclosure Baylor College of Medicine has filed for intellectual property protection on behalf of authors CD, PN, and AM on material related to the device in this manuscript. Remaining authors declare no competing interests.
Figures




References
Manuscript References
-
- Naghavi M. et al. Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. The Lancet 404, 1199–1226 (2024). 10.1016/S0140-6736(24)01867-1 - DOI
-
- Pirnay J.-P. et al. Personalized bacteriophage therapy outcomes for 100 consecutive cases: a multicentre, multinational, retrospective observational study. Nature Microbiology 9, 1434–1453 (2024). 10.1038/s41564-024-01705-x - DOI
-
- Hutinet G. et al. 7-Deazaguanine modifications protect phage DNA from host restriction systems. Nature Communications 10, 5442 (2019). 10.1038/s41467-019-13384-y - DOI
Methods References*
-
- Apprill A., McNally S. P., Parsons R. J. & Weber L.
-
- Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K. & Schloss P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79, 5112–5120 (2013). 10.1128/AEM.01043-13 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
