This is a preprint.
Genome-wide screen in Mycobacterium tuberculosis infected macrophages reveals innate regulation of antibacterial mediators by IRF2
- PMID: 41256720
- PMCID: PMC12621954
- DOI: 10.1101/2025.09.26.678671
Genome-wide screen in Mycobacterium tuberculosis infected macrophages reveals innate regulation of antibacterial mediators by IRF2
Abstract
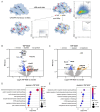
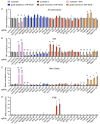
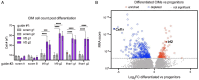
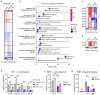
Controlling Mycobacterium tuberculosis (Mtb) infection requires a precisely balanced host inflammatory response. Too little inflammation leads to uncontrolled bacterial growth but exacerbated inflammation, activated by mediators such as TNF and type I IFN, inhibits effective antibacterial responses. How these immunopathological states are established is unknown. Deeper understanding of the pathways elicited upon initial Mtb infection of the host macrophage may reveal vital regulatory mechanisms that govern the subsequent inflammatory environment and ultimate resolution of infection. To elucidate these early regulators of inflammation, we performed a genome-wide CRISPR knockout screen in macrophages to identify genes that influence the induction of TNF and iNOS upon infection with Mtb. The resulting dataset is a valuable resource that includes genes representing a wide range of unexpected regulatory mechanisms that control cytokine responses to Mtb and also cell-intrinsic resistance to infection by the bacterial pathogen Listeria monocytogenes. We show that type I IFN signaling enhances TNF production early after infection, and IRF2 acts to inhibit induction of the antibacterial state of macrophages. Our data support a model in which early production of type I IFN in response to bacterial infection serves to increase innate antibacterial resistance during the earliest stages of infection.
Figures



References
-
- Boneca I.G., Dussurget O., Cabanes D., Nahori M.-A., Sousa S., Lecuit M., Psylinakis E., Bouriotis V., Hugot J.-P., Giovannini M., Coyle A., Bertin J., Namane A., Rousselle J.-C., Cayet N., Prévost M.-C., Balloy V., Chignard M., Philpott D.J., Cossart P., and Girardin S.E.. 2007. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proceedings of the National Academy of Sciences. 104:997–1002. doi: 10.1073/pnas.0609672104. - DOI
-
- Bulua A.C., Simon A., Maddipati R., Pelletier M., Park H., Kim K.-Y., Sack M.N., Kastner D.L., and Siegel R.M.. 2011. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J Exp Med. 208:519–533. doi: 10.1084/jem.20102049. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
