Automated hypoxia and apnea identification for neonates via enhanced respiratory signal modeling with deep learning
- PMID: 41257997
- PMCID: PMC12630711
- DOI: 10.1038/s41598-025-24783-1
Automated hypoxia and apnea identification for neonates via enhanced respiratory signal modeling with deep learning
Abstract
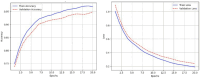
Neonatal respiratory monitoring is crucial for assessing breathing patterns, but the lack of real-time clinical data limits the development of machine learning (ML) models. This study provides a synthetic signal generation framework to replicate infant respiratory cycles with physiological fidelity. The dataset simulates normal and pathological breathing patterns such as apnea, hypoxia, and periodic breathing, including Gaussian noise and exponential functions, to maintain biological realism. A feature extraction pipeline was created to examine time- and frequency-domain characteristics, enabling the categorization of respiratory states using Convolutional Neural Networks (CNNs), CNN - BiLSTM and Random Forests. The CNN-BiLSTM model achieved the highest classification accuracy of 96.16%, outperforming the standalone CNN and RF models. The results illustrate the possibility of synthetic neonatal data for ML-based respiratory distress assessment. This architecture can be further extended for hardware implementation using e-textile-based respiratory monitoring. Real neonatal dataset integration and clinical validation of ML-DL models will be the main goals of future research to improve their robustness and applicability.
Keywords: Apnea and hypoxia classification; Machine learning in NICU; Neonatal respiratory monitoring; Synthetic biomedical signal generation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures















References
-
- Chen, W. et al. Design of an integrated sensor platform for vital sign monitoring of newborn infants at neonatal intensive care unit. J. Healthc. Eng.1 (4), 535–554. 10.1260/2040-2295.1.4.535 (2010).
MeSH terms
LinkOut - more resources
Full Text Sources

