Systematic benchmarking of imaging spatial transcriptomics platforms in FFPE tissues
- PMID: 41266321
- PMCID: PMC12635344
- DOI: 10.1038/s41467-025-64990-y
Systematic benchmarking of imaging spatial transcriptomics platforms in FFPE tissues
Abstract
Emerging imaging spatial transcriptomics (iST) platforms and coupled analytical methods can recover cell-to-cell interactions, groups of spatially covarying genes, and gene signatures associated with pathological features, and are thus particularly well-suited for applications in formalin fixed paraffin embedded (FFPE) tissues. Here, we benchmark the performance of three commercial iST platforms-10X Xenium, Vizgen MERSCOPE, and Nanostring CosMx-on serial sections from tissue microarrays (TMAs) containing 17 tumor and 16 normal tissue types for both relative technical and biological performance. On matched genes, we find that Xenium consistently generates higher transcript counts per gene without sacrificing specificity. Xenium and CosMx measure RNA transcripts in concordance with orthogonal single-cell transcriptomics. All three platforms can perform spatially resolved cell typing with varying degrees of sub-clustering capabilities, with Xenium and CosMx finding slightly more clusters than MERSCOPE, albeit with different false discovery rates and cell segmentation error frequencies. Taken together, our analyses provide a comprehensive benchmark to guide the choice of iST method as researchers design studies with precious samples in this rapidly evolving field.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.W. receives research support from Merck Sharp & Dohme, 10X Genomics, and research collaboration agreement with NanoString. Consumables used in this study from both companies were purchased at full price. The remaining authors declare no competing interests.
Figures
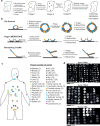

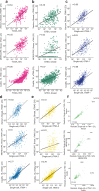
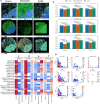

Update of
-
Systematic benchmarking of imaging spatial transcriptomics platforms in FFPE tissues.bioRxiv [Preprint]. 2023 Dec 19:2023.12.07.570603. doi: 10.1101/2023.12.07.570603. bioRxiv. 2023. Update in: Nat Commun. 2025 Nov 20;16(1):10215. doi: 10.1038/s41467-025-64990-y. PMID: 38106230 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

