To be or not to be a protein coding mutation, that's the question!
- PMID: 41278537
- PMCID: PMC12634407
- DOI: 10.1093/nargab/lqaf168
To be or not to be a protein coding mutation, that's the question!
Abstract
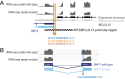
Accurate annotation of genetic variants-distinguishing whether they affect protein-coding or noncoding genomic regions-is crucial for evaluating their potential role in disease development. Prominent examples have been identified of variants that for many years had been considered to be coding missense or synonymous mutations targeting one gene, and that recently turned out to be noncoding variants, sometimes even modulating a shared regulatory region of multiple genes. These errors were caused by annotating to a canonical reference transcript, whereas an alternative transcript was in reality expressed in respect to which the mutations have a different annotation. Unfortunately, this practice of annotating genetic variants to a reference transcript, without verifying whether this transcript is expressed or whether the mutation causes a change of expressed transcript, is still widespread. However, the implementation of RNA sequencing and availability of these data in online portals allow to verify expressed transcripts in relevant tissues. Integration of DNA- and RNA-sequencing data, in which detected DNA mutations are annotated in respect to the transcripts that are expressed in the corresponding tissue or disease sample as detected by RNA sequencing, avoids misinterpretation of noncoding variants as coding and vice versa, thereby improving the functional interpretation of genetic variants.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
-
- Carlin JL. Mutations are the raw materials of evolution. Nat Educ Knowl. 2011;3:10.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

