This is a preprint.
Macrodomain ADP-ribose binding but not ADP-ribosylhydrolase activity is critical for chikungunya virus infection of Aedes mosquitoes
- PMID: 41279946
- PMCID: PMC12632316
- DOI: 10.1101/2025.10.09.681498
Macrodomain ADP-ribose binding but not ADP-ribosylhydrolase activity is critical for chikungunya virus infection of Aedes mosquitoes
Abstract
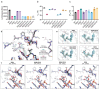
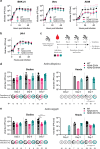
Viral macrodomains are promising antiviral targets that counteract host ADP-ribosylation-mediated antiviral responses in mammals. However, their role in dual-host viruses within the mosquito vector is largely unknown. Here, we investigated the role of the chikungunya virus (CHIKV) macrodomain by mutating the active site asparagine 24 (N24). In both mammalian and mosquito cell lines, these enzymes rapidly acquired compensatory mutations at aspartate 31 (D31). We show that while N24 mutations abolish ADP-ribosylhydrolase catalytic activity and reduce folding stability, ADP-ribose binding remains intact. Furthermore, the D31 compensatory mutations do not markedly rescue catalytic activity or folding stability. Structures of the compensatory mutant macrodomains suggest the importance of ADP-ribose binding, rather than ADP-ribosylhydrolase catalysis as the selective pressure driving their accumulation. In mammalian cells, viral mutants bearing the catalytic and compensatory mutations replicated less efficiently than wild-type virus in interferon-competent cell lines. However, their replication remained unaffected in mosquito cells. In Aedes mosquitoes, macrodomain mutations had disparate impacts, either reducing or enhancing infectivity and transmission depending on the specific mutation and viral lineage. These findings emphasize that viral macrodomain function is complex and host-dependent, highlighting the need for multi-host understanding to develop effective antivirals.
Conflict of interest statement
Declaration of interests A.A. is a co-founder of Tango Therapeutics, Azkarra Therapeutics and Kytarro; a member of the board of Cytomx, Ovibio Corporation, Cambridge Science Corporation; a member of the scientific advisory board of Genentech, GLAdiator, Circle, Bluestar/Clearnote Health, Earli, Ambagon, Phoenix Molecular Designs, Yingli/280Bio, Trial Library, ORIC and HAP10; a consultant for ProLynx, Next RNA and Novartis; receives research support from SPARC; and holds patents on the use of PARP inhibitors held jointly with AstraZeneca from which he has benefited financially (and may do so in the future). J.S.F. is a consultant to, shareholder of, and receives sponsored research support from Relay Therapeutics and is a compensated member of the SAB of Vilya Therapeutics. The other authors declare no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
