This is a preprint.
CEBPB, C19MC, and Defective Autophagy Drive Novel Podosomal Belt to Macropinocytosis Transition, Lipid Accumulation, and HBV A-to-I RNA-editing
- PMID: 41282072
- PMCID: PMC12636704
- DOI: 10.21203/rs.3.rs-6805130/v1
CEBPB, C19MC, and Defective Autophagy Drive Novel Podosomal Belt to Macropinocytosis Transition, Lipid Accumulation, and HBV A-to-I RNA-editing
Abstract
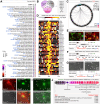
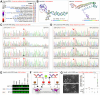
Obesity and neurodegeneration are clinically associated diseases with defective autophagy. However, the genetic, biological, and metabolic underpinnings connecting these diseases are not well-understood. Here we identified a Mitochondria obesity/neurodegeneration (M on ) gene-signature that is shared between obesity, and neurodegenerative diseases. We demonstrate that, CEBPB elevates M on -gene-signature, to form podosomal belts, and enhance ROS production. Inhibiting autophagy collapses podosomal-belts through macropinocytosis to accumulate vacuoles, lipid-droplets, nuclear Notch-1 (nNICD), DEPTOR, and HBV-polymerase mRNAs. Conversely, hemin counteracts these events and suppresses DEPTOR and HBV-polymerase mRNAs by A-to-I-RNA-editing and nonsense-mediated decay. Furthermore, we CRISPR-engineered the antiviral chromosome-19 miRNA cluster (C19MC) to demonstrate that C19MC-miRNAs augment CEBPB, M on -gene-signature, ROS, and recapitulate CEBPB-driven phenotypes, in response to autophagy inhibition. Hemin, or a γ-Secretase inhibitor counteract these phenotypes in CRISPR-C19MC-engineered cells. Therefore, a CEBPB and C19MC-driven M on -gene-signature regulates the podosomal belt, lipid droplet, HBV, and DEPTOR mRNA dynamics to genetically link obesity, and neurodegeneration at the cellular level.
Conflict of interest statement
Competing interest statement
The authors G.G.J., and A.S.B. have a provisional patent application pending for the RNA probe Cy5-CSi000A, and CRISPR-guides-3 and 4 that are used to generate ZNF331-C19MC fusion genetic engineering. A.S.B. has advisory board relationships with Deciphera. The remaining authors have no other conflicts of interests to declare. All authors agree to take individual responsibility and accountability in the event if found any undeclared conflicts of interest(s) that existed before or at the time of manuscript submission.
Figures






References
-
- Sargent J. Neurodegenerative disease: Balancing BMI--rethinking the relationships between obesity, ageing and risk of dementia. Nat Rev Endocrinol 11, 315 (2015). - PubMed
-
- Procaccini C. et al. Role of metabolism in neurodegenerative disorders. Metabolism 65, 1376–1390 (2016). - PubMed
-
- Knopman D.S. et al. Alzheimer disease. Nature Reviews Disease Primers 7, 33 (2021).
Publication types
LinkOut - more resources
Full Text Sources

