This is a preprint.
Melanoma to rhabdomyosarcoma plasticity in the setting of immunotherapy
- PMID: 41282820
- PMCID: PMC12637767
- DOI: 10.1101/2025.10.31.25338685
Melanoma to rhabdomyosarcoma plasticity in the setting of immunotherapy
Abstract
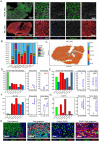
Acquired resistance to immune checkpoint inhibitors (ICIs) remains a significant challenge in the treatment of metastatic melanoma. Phenotypic plasticity, such as dedifferentiation and transdifferentiation, is an increasingly recognized mechanism for treatment resistance. We present a case of a man in his 70s with metastatic melanoma who experienced progression through sequential treatments including pembrolizumab in combination with the HDAC inhibitor entinostat, and ipilimumab. During treatment a histologically distinct pleomorphic rhabdomyosarcoma (RMS) emerged at metastatic sites. Longitudinally acquired tumor samples representing both phenotypes were analyzed using whole-exome sequencing (WES), RNA sequencing (RNA-seq) and high-plex tissue imaging (spatial proteomics). WES revealed driver mutations (e.g. NRAS, NF1) and loss-of-heterozygosity (LOH) shared between phenotypes indicating a common ancestral clone. Phylogenetic analysis demonstrated an early divergence of the phenotypes, with each later acquiring unique mutations. RNA-seq showed mutually exclusive expression of lineage-specific markers as well as epithelial-mesenchymal transition and myogenic gene set enrichment in the RMS samples. High-plex imaging identified distinct tumor microenvironments, with RMS lesions enriched in CD163+ macrophages.
Figures


References
-
- Chu X. et al. Neuroendocrine transformation from EGFR/ALK-wild type or TKI-naïve non-small cell lung cancer: An under-recognized phenomenon. Lung Cancer 169, 22–30 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
