Elevated expression of SIGLEC8 in skin cutaneous melanoma predicts better prognosis associated with hematopoietic cell lineage pathway
- PMID: 41288776
- PMCID: PMC12647512
- DOI: 10.1007/s12672-025-03935-9
Elevated expression of SIGLEC8 in skin cutaneous melanoma predicts better prognosis associated with hematopoietic cell lineage pathway
Abstract
Background: Cutaneous melanoma (SKCM) is becoming more common. Current prognostic markers-TNM stage, BRAF mutation status, and tumor mutational burden (TMB)-do not offer precise predictions. Recent studies point to a key role for the tumor microenvironment. To date, we do not know how SIGLEC8 (a sialic acid-binding immunoglobulin‑like lectin) affects immune regulation or prognosis in SKCM.
Methods: We used logistic regression to link SIGLEC8 expression with clinical and pathological features. We then applied Cox regression and Kaplan‑Meier survival analysis to assess how these features relate to overall survival (OS). For differential genes, we performed Gene Ontology (GO) and KEGG enrichment tests by combining fold‑change thresholds with hypergeometric statistics. Finally, we ran Pearson and Spearman correlations to study the relationship between SIGLEC8 and immune‑infiltration markers.
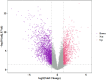
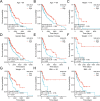
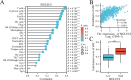
Results: From TCGA, we selected SKCM cases that had both complete clinical records and transcriptome data. We excluded 8.2% of samples due to missing information; this did not introduce any bias in age or sex. Patients with low SIGLEC8 levels tended to have T4 tumors, Breslow depth over 3 mm, ulceration, and more advanced disease (p < 0.05). High SIGLEC8 expression was linked to better OS (p < 0.001) in Cox analysis. This prognostic value held true across subgroups defined by age, gender, BMI, and stage (all p < 0.05). Differential expression analysis highlighted three immune genes-FCER2, CR2, and MS4A1-as strongly correlated with SIGLEC8. These findings point to a role in B‑cell and CD8⁺ T‑cell pathways within the hematopoietic lineage.
Conclusions: High SIGLEC8 expression predicts longer survival in SKCM. We propose that SIGLEC8 boosts antitumor immunity by influencing B‑cell and T‑cell pathways (including FCER2, CR2, MS4A1, and CD8A/B) in the tumor microenvironment. SIGLEC8 thus shows promise as a low‑cost prognostic biomarker and a potential therapeutic target. Table S1 shows cancer-type-specific prognostic effects for SIGLEC8: in SKCM higher SIGLEC8 expression associates with improved survival, whereas prognostic directionality varies across other tumor types. A pan-cancer comparison revealed cancer-type-specific prognostic effects for SIGLEC8: in SKCM higher SIGLEC8 expression associates with improved survival, whereas prognostic directionality varies across other tumor types.
Keywords: Prognostic biomarker; SIGLEC8; SKCM; TCGA; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. This study utilized publicly available, de-identified data from The Cancer Genome Atlas (TCGA). Since no new human participants were recruited and no identifiable private information was accessed or generated, ethical approval for this specific retrospective analysis was not required. The original TCGA study protocols received approval from the relevant Institutional Review Boards (IRBs). Written informed consent was obtained from all participants in the original TCGA study. Given the exclusive use of de-identified, publicly available data from TCGA in this secondary analysis, obtaining new consent to participate was not required. Consent for publication: Informed consent for publication was obtained from all participants during their enrollment in the original TCGA study. Given the exclusive use of de-identified, aggregated data in this analysis, separate consent for publication is not required. Competing interests: The authors declare no competing interests.
Figures







References
-
- Eggermont AM, Spatz A, Robert C. Cutaneous melanoma. Lancet. 2014;383(9919):816–827. - PubMed
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. - PubMed
-
- Whiteman DC, Green AC, Olsen CM. The growing burden of invasive melanoma: projections of incidence rates and numbers of new cases in six susceptible populations through 2031. J Invest Dermatol. 2016;136(6):1161–71. - PubMed
Grants and funding
LinkOut - more resources
Research Materials
Miscellaneous
