Unraveling the genetic architecture of anti-nutritional factors in soybean (Glycine max.) for nutritional enhancement
- PMID: 41315536
- PMCID: PMC12663474
- DOI: 10.1038/s41598-025-27132-4
Unraveling the genetic architecture of anti-nutritional factors in soybean (Glycine max.) for nutritional enhancement
Abstract
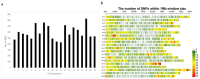
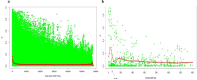
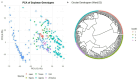
Anti-nutritional factors (ANFs) can reduce nutrient bioavailability for monogastric animals. Therefore, this study aimed to understand the genetic architecture underlying ANF accumulation in soybean. Diversity arrays technology and a spectrophotometric method were employed to generate genotypic and phenotypic data, respectively, and gene mining was performed within 100-kb genomic window. A significant difference was found regarding ANFs content in the genotypes (p < 0.001). Significant SNP markers for phytate were identified on chromosomes 3, 4, 13, and 20 by FarmCPU, and for total trypsin inhibitors (TTI) on 6, 12, and 14 by CMLM models, whereas mrMLM model detected markers on chromosome 3, 12 and 15 for phytate, 4, 9, 13, 17 and 18 for TTI. Genes associated with phytate content include Glyma.03G001600, Glyma.04G194600, Glyma.13G128200, Glyma.20G118700, Glyma.14G213400, and Glyma.16G126400. For TTI, the genes are Glyma.06G074700, Glyma.12G241600, Glyma.14G176700, Glyma.13G052700, and Glyma.18G050400. These genes are primarily linked to plant defense and substrate interactions. Most promising SNP markers for marker-assisted selection aimed at reducing phytate levels include Soy_3_218818 (218,818 bp), Soy_3_241209 (241,209 bp), Soy_4_45462019 (45,462,019 bp), Soy_14_48672982 (48,672,982 bp), and Soy_6_5695090 (5,695,090 bp). For TTI, key markers include Soy_14_43649238 (43,649,238 bp), Soy_12_41339023 (41,339,023 bp), Soy_18_4301721 (4,301,721 bp), and Soy_13_14029215 (14,029,215 bp). These findings offer a valuable foundation for marker-assisted breeding aimed at improving soybean nutritional quality.
Keywords: GWAS; Phytate; SNP markers; Soybean; Trypsin inhibitors.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval and consent to participate: The seeds used in the study are owned by the Makerere University Centre for Soybean Improvement and Development (MAKCSID) led by Senior Soybean Breeder in Uganda, Prof. Phinehas Tukamuhabwa. Therefore, the collection of the seeds used in the study complies with local or national guidelines with no need for further affirmation. Consent for publication: All authors have read and agreed to the published version of the manuscript.
Figures



References
-
- Gibbs, B. F., Zougman, A., Masse, R. & Mulligan, C. Production and characterization of bioactive peptides from soy hydrolysate and soy-fermented food. Food Res. Int.37, 123–131 (2004).
-
- Murithi, H. M., Beed, F., Tukamuhabwa, P., Thomma, B. P. H. J. & Joosten, M. H. A. J. Soybean production in eastern and southern Africa and threat of yield loss due to soybean rust caused by Phakopsora pachyrhizi. Plant Pathol.65, 176–188 (2016).
-
- Rui, X. et al. Optimization of soy solid-state fermentation with selected lactic acid bacteria and the effect on the anti-nutritional components. J. Food Process. Preserv.10.1111/jfpp.13290 (2017).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

