Discovery of a Novel Non-Nucleoside Inhibitor of RNA-Dependent RNA Polymerase Against Dengue Virus
- PMID: 41328325
- PMCID: PMC12664905
- DOI: 10.1002/mco2.70514
Discovery of a Novel Non-Nucleoside Inhibitor of RNA-Dependent RNA Polymerase Against Dengue Virus
Abstract
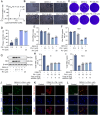
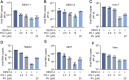
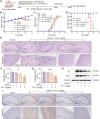
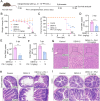
Dengue virus (DENV) is an acute infectious pathogen worldwide, for which no effective therapeutics are available. The RNA-dependent RNA polymerase (RdRp) displays an important role during DENV replication and is therefore a promising target in the development of antiviral drugs. However, there are still no clinically approved RdRp inhibitors available. In this study, we identified a natural small molecule, 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside (PN-1), using a surface plasmon resonance-based screening assay. Biochemical and structural analyses revealed that PN-1 selectively targets the RdRp of DENV NS5 protein by covalently modifying residues Glu255, Met387, Glu479, and Ala507. Mechanistic studies involving tryptophan scanning and hydrogen-deuterium exchange mass spectrometry revealed that PN-1 binding regulates RdRp conformational transitions. This allosteric mechanism leads to suppression of enzymatic activity and inhibition of DENV replication. Consequently, PN-1 exhibited potent antiviral activity across various cell lines and conferred significant protection in both ICR suckling and AG129 mouse models. Taken together, our results show that PN-1 functions as a novel non-nucleoside inhibitor to suppress DENV replication by targeting RdRp. These findings highlight PN-1 as a promising anti-DENV lead compound, while revealing conserved RdRp residues as actionable targets for rational drug design.
Keywords: AG129 mice; ICR suckling; RNA‐dependent RNA polymerase; dengue virus; target.
© 2025 The Author(s). MedComm published by Sichuan International Medical Exchange & Promotion Association (SCIMEA) and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Simmons C. P., Farrar J. J., Nguyen V. V., and Wills B., “Dengue,” New England Journal of Medicine 366 (2012): 1423–1432. - PubMed
-
- World Health Organization , "Dengue and Severe Dengue," accessed April 23, 2024, https://www.who.int/news‐room/fact‐sheets/detail/dengue‐and‐severe‐dengue.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
