Cell-type specific single-cell signatures reveal nephrotoxic drug effects
- PMID: 41334180
- PMCID: PMC12666064
- DOI: 10.1016/j.csbj.2025.11.012
Cell-type specific single-cell signatures reveal nephrotoxic drug effects
Abstract
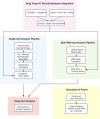
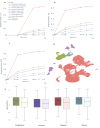
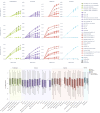
Drug-induced acute kidney injury (AKI) affects about 20 % of hospitalized AKI patients, a significant contributor to morbidity and mortality. The lack of understanding of the kidney system and functioning of nephrotoxic drugs contributes to hospital-acquired AKI cases. AKI is difficult to predict because of its complex injury mechanism and the numerous pathways through which it manifests. Traditional toxicity biomarkers, like elevated creatinine levels, detect AKI only after significant kidney injury has occurred. Concurrently, advancements in single cell RNA sequencing (scRNAseq) have improved our ability to map cellular heterogeneity within tissues, potentially enabling the study of drug effects at a single cell level. We hypothesized that only particular subtypes of kidney cells may be responsible for observed nephrotoxicity and explain prediction challenges. To test this, we generated cellular response scores for 32 kidney cell types from the Human Cell Atlas and estimated drug effects. We identified significant expression differences in 6 cell types (e.g. Indistinct intercalated cell p = 0.009, Epithelial Progenitor cell, p = 0.04). We also developed an XGBoost model that achieved an AUROC of 0.6 on an external test set, across different kidney cell populations - a significant improvement over using traditional bulk RNA sequencing alone. The single-cell transcriptomic signatures we identified potentially reveal unexplained molecular mechanisms of nephrotoxicity. This work provides both a reproducible computational framework and curated dataset available at https://doi.org/10.5281/zenodo.15724290 to the research community.
Keywords: Nephrotoxicity; Single cell RNAseq; Single cell signatures.
© 2025 The Authors.
Figures

Update of
-
Cell-Type Specific Single-Cell Signatures Reveal Nephrotoxic Drug Affects.bioRxiv [Preprint]. 2025 Jun 22:2025.06.17.660070. doi: 10.1101/2025.06.17.660070. bioRxiv. 2025. Update in: Comput Struct Biotechnol J. 2025 Nov 06;27:5148-5158. doi: 10.1016/j.csbj.2025.11.012. PMID: 40667011 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
