Unraveling forensic timelines using molecular markers in Phormia regina maggots
- PMID: 41343428
- PMCID: PMC12677486
- DOI: 10.1371/journal.pgen.1011948
Unraveling forensic timelines using molecular markers in Phormia regina maggots
Abstract
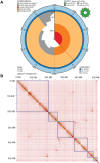
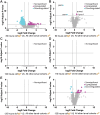
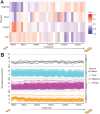
In the medico-legal application of forensic entomology, estimating the time of death is critical and traditionally relies on changes in observable traits of carrion feeding insect larvae. Traits such as size, weight, and morphology can be used to predict the insect specimen age and help define the minimum time since death. The blowfly Phormia regina Meigen (Diptera: Calliphoridae) is a key forensic insect, yet age estimation for older maggots in this and other carrion-feeding species is particularly challenging due to the limited morphological changes in the late-stage larvae. To enhance age-estimation precision, we employed transcriptomic profiling on blowfly maggots, aiming to identify genes as markers for time of death estimation. Our study characterized maggot development, reinforcing that weight and behavior cannot precisely determine age between 100 and 130 hours at 27.5 °C. We built a chromosomal scale annotated genome, establishing a reliable database for uncovering transcriptomic signatures during larval development. Applying differential gene expression analyses, weighted gene co-expression network analysis, and the generalized linear model, we identified nine candidate genes (y5078, y5076, agt2, ech1, dhb4, asm, gabd, acohc, ivd) that delineate the age of otherwise indeterminate maggots. This research introduces a molecular approach to address a longstanding problem in forensic entomology and promises to increase precision in determining the time of death at a crime scene.
Copyright: © 2025 Lin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Update of
-
Unraveling forensic timelines using molecular markers in Phormia regina maggots.bioRxiv [Preprint]. 2025 May 7:2025.04.30.651557. doi: 10.1101/2025.04.30.651557. bioRxiv. 2025. Update in: PLoS Genet. 2025 Dec 4;21(12):e1011948. doi: 10.1371/journal.pgen.1011948. PMID: 40654918 Free PMC article. Updated. Preprint.
References
-
- Kasper J. Examinations of decomposition-processes and the olfactory sense of the necrophagous fly Lucilia Caesar (L.) (Diptera: Calliphoridae) in relationship to the fly’s physiological state. 2013.
-
- Wells JD, LaMotte LR. Estimating the postmortem interval. In: Byrd JH, Tomberlin JK. Forensic Entomology: Utility of Arthropods in Legal Investigations. 3rd ed. CRC Press. 2020. 213–24.
-
- Walden SJ, Evans SL, Mulville J, Wilson K, Board S. Progressive dehydration in decomposing bone: a potential tool for forensic anthropology. J Therm Anal Calorim. 2021;143:3517–24.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

