Progressively exploring and assessing the prognosis of bladder urothelial cancer based on the microenvironment through the integration of multiple databases
- PMID: 41346448
- PMCID: PMC12672317
- DOI: 10.3389/fmolb.2025.1702311
Progressively exploring and assessing the prognosis of bladder urothelial cancer based on the microenvironment through the integration of multiple databases
Abstract
Background: The heterogeneous prognosis of bladder urothelial carcinoma (BLCA) remains a significant clinical challenge. A multi-factor prognostic model is essential for BLCA, as it not only assesses tumor progression and elucidates underlying molecular mechanisms but also paves the way for timely treatment adjustments and improved clinical decision-making.
Methods: Using R software, we performed immunophenotyping on multiple BLCA cohorts from the GEO database to identify shared immune signatures. Simultaneously, we identified BLCA prognosis-associated genes by analyzing TCGA data. Prognostic genes were further refined via LASSO regression, allowing BLCA patients to be stratified into high- and low-risk groups based on their expression patterns. Quantitative PCR (qPCR) was used to validate gene expression in tumor and matched normal tissues. Finally, we integrated clinical data to construct a prognostic model.
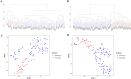
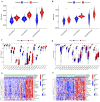
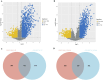
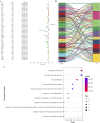
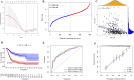
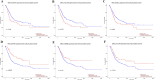
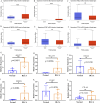
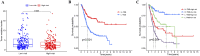
Results: The GSE31684 and GSE48276 cohorts were divided into high immunity (Immunity_H) and low immunity (Immunity_L) groups, and there were significant microenvironment differences between the Immunity_H and Immunity_L of the two cohorts, and there were many common differentially expressed genes (DEGs) between different immune subtypes of the two cohorts, which were mainly involved in immune-related biological processes. In addition, patients in the high-risk BLCA group exhibited significantly worse prognosis than those in the low-risk group. qPCR analysis confirmed that the expression levels of the risk-stratification genes were significantly different between BLCA tumors and matched adjacent normal tissues. The integrated analysis of tumor mutation burden (TMB) and our risk stratification revealed that patients with low-risk scores and high TMB exhibited the most favorable prognosis. Furthermore, the risk score was validated as an independent prognostic factor through both univariate and multivariate Cox regression analyses. Consequently, we constructed a nomogram that incorporates these findings to assist clinicians in prognostic assessment for BLCA patients.
Conclusion: Given that the tumor microenvironment significantly influences BLCA prognosis, our finding that risk stratification serves as an independent prognostic indicator underscores the clinical relevance of our model. This stratification strategy has the potential to improve prognostic assessment and inform personalized treatment planning for BLCA patients.
Keywords: bladder urothelial carcinoma (BLCA); immune subtypes; microenvironment; prognosis; risk stratification.
Copyright © 2025 Zou, Li, Peng, Gu and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

