F9 missense variant hot spots associated with qualitative protein defects causing hemophilia B
- PMID: 41356341
- PMCID: PMC12680096
- DOI: 10.1016/j.bvth.2025.100095
F9 missense variant hot spots associated with qualitative protein defects causing hemophilia B
Abstract
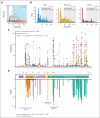
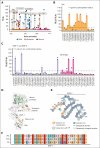
Hemophilia B, a rare bleeding disorder, is caused by genetic variations in F9. Although quantitative factor IX (FIX) deficiencies are successfully treated by protein replacement therapy, qualitative defects may result in dysfunctional proteoforms in the circulation, potentially interfering with prophylactically or therapeutically administered recombinant FIX (rFIX) concentrates. To delineate the F9 missense variants associated with qualitative defects, we integrated genotype and phenotype from the European Association for Haemophilia and Allied Disorders F9 Coagulation Factor Variant Database. Of the 663 patients for whom activity (FIX:Act) and antigen (FIX:Ag) data were available, 40% of patients with severe hemophilia (n = 248), 50% of patients with moderate hemophilia (n = 244), and 47% of patients with mild hemophilia (n = 171) had cross-reactive material defined as FIX:Ag ≥40%. Variants associated with qualitative defects were predominantly localized (1) at proteolytic sites for FIX processing and activation, (2) within exosite II of the serine protease domain, and (3) at calcium ion coordinating residues within the Gla/EGF-1 domain of the FIX light chain. To study the effect of dysfunctional FIX proteoforms on thrombin generation in the presence of rFIX, we investigated 2 individuals with hemophilia B harboring a variant of unknown significance with an unexplained bleeding phenotype despite prophylaxis. Ex vivo therapeutic monitoring using patient plasma supplemented with rFIX concentrates, bypassing agent or emicizumab, enabled head-to-head comparison and revealed limited normalization of the prolonged initiation phase in coagulation. Alignment of information on genotype with functional proteotype may clarify the heterogeneity in bleeding phenotype and treatment response and provide a stepping stone for personalized care.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: T.T.v.D. received research grants from The Society for the Advancement of Science, Medicine and Surgery (Het Genootschap ter bevordering van Natuur- Genees- en Heelkunde); and the Professor Heimburger award from Commonwealth Serum Laboratories Behring. The institution of K.F. has received unrestricted research grants from Commonwealth Serum Laboratories Behring, Sobi, and Novo Nordisk; and consultancy fees from F. Hoffmann-La Roche, Sanofi, Sobi, and Novo Nordisk. J.C.M.M. received consultancy fees from Alveron Pharma and Synapse Research Institute. The remaining authors declare no competing financial interests.
Figures





References
-
- Zhang H, Xin M, Lin L, Chen C, Balestra D, Ding Q. Pleiotropic effects of different exonic nucleotide changes at the same position contribute to hemophilia B phenotypic variation. J Thromb Haemost. 2024;22(4):975–989. - PubMed
-
- Srivastava A, Santagostino E, Dougall A, et al. WFH guidelines for the management of hemophilia, 3rd edition. Haemophilia. 2020;26(suppl 6):1–158. - PubMed
-
- Verhagen MJA, van Balen EC, Blijlevens NMA, et al. Patients with moderate hemophilia A and B with a severe bleeding phenotype have an increased burden of disease. J Thromb Haemost. 2024;22(1):152–162. - PubMed
-
- Castaman G, Jimenez-Yuste V, Gouw S, D’Oiron R. Outcomes and outcome measures. Haemophilia. 2024;30(suppl 3):112–119. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
