Development of a senescence-related lncRNA signature in endometrial cancer based on multiple machine learning models
- PMID: 41384040
- PMCID: PMC12694935
- DOI: 10.3389/fgene.2025.1687922
Development of a senescence-related lncRNA signature in endometrial cancer based on multiple machine learning models
Abstract
Background: Senescence-related lncRNAs (srlncRNA) mediate carcinogenesis in various malignancies. However, its roles in endometrial cancer (EC) remain unknown. Our research aims to construct a predictive srlncRNA model with prognostic and therapeutic significance in EC.
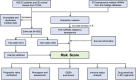
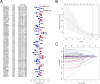
Methods: We first downloaded the gene expression and medical information from the TCGA, as well as senescence-related lncRNAs (srlncRNAs) from the CellAge databases. Then, a co-expression network of cell senescence-related mRNA-lncRNA was explored with R. Subsequently, we performed Cox and Lasso regression and machine learning analysis to identify srlncRNAs related to the prognosis of EC and built a predictive model. Continually, we drew a nomogram to improve its ability to predict prognosis. Further, GSEA was used to explore potential mechanisms. Differences in TME, immune infiltrating cells, and checkpoints of the two risk groups were compared using GSEA and CIBERSORT. Finally, the drug sensitivity of patient-derived tumor organoids (PDOs) was investigated.
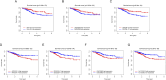
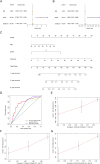
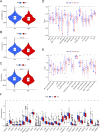
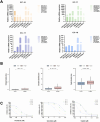
Results: We first built a prognostic model based on seven srlncRNAs (AL121906.2, AP002761.4, BX322234.1, LINC00662, LINC00908, VIM-AS1, and ZNF236-DT). The model, which was screened by machine learning, functioned well in three sets with good stability and accuracy. Furthermore, the nomogram based on age, grade, and risk scores could precisely predict the prognosis of EC patients. The AUC of risk scores was highest compared to other clinical parameters (AUC risk score = 0.769, AUC age = 0.615, and AUC grade = 0.681). This srlncRNAs were enriched in the cell cycle, certain malignant tumors, and cancer-associated regulatory pathways. Afterward, low-risk EC patients had more immune-infiltrating cells and may benefit from anti-PD-1 and anti-CTLA4 treatment. Paclitaxel, gemcitabine, and cisplatin (all p < 0.05) may be more useful in EC patients with high expression of targeted srlncRNAs in the GDSC database. The levels of targeted srlncRNAs and drug sensitivity varied significantly among different EC PDOs. The EC-18 PDO was more resistant to three drugs, which aligned with clinical observation.
Conclusion: The srlncRNA signature (AL121906.2, AP002761.4, BX322234.1, LINC00662, LINC00908, VIM-AS1, and ZNF236-DT) could guide prognosis prediction and treatment choices for EC patients.
Keywords: TCGA; cell senescence; endometrial cancer; immunotherapy; patient-derived tumor organoids; prognosis; signature.
Copyright © 2025 Lin, Lei, Li, Jiang, Jiang, Guo, Cai, Ye and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Chen H.-A., Ho Y.-J., Mezzadra R., Adrover J. M., Smolkin R., Zhu C., et al. (2023). Senescence rewires microenvironment sensing to facilitate Antitumor immunity. Cancer Discov. 13, 432–453. 10.1158/2159-8290.CD-22-0528 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

