Histone methyltransferase DOT1L maintains cell state and restricts cytotoxic potential of CD8 T cells
- PMID: 41385642
- PMCID: PMC12700201
- DOI: 10.1126/sciadv.adw1289
Histone methyltransferase DOT1L maintains cell state and restricts cytotoxic potential of CD8 T cells
Abstract
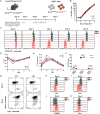
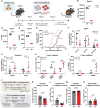
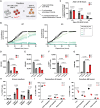
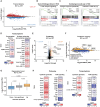
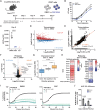
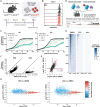
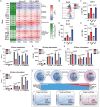
The histone methyltransferase DOT1L is emerging as a central epigenetic regulator in immune cells. Loss of DOT1L during development of CD8 T cells in vivo leads to gain of memory characteristics but has also been reported to compromise CD8 T cell viability and antitumor reactivity. Here, we determined the cell-intrinsic role of DOT1L in mature mouse CD8 T cells. After conditional deletion of Dot1L in vitro, CD8 T cells retained in vivo proliferative capacity and antitumor reactivity. Moreover, Dot1L knockout CD8 T cells showed increased antigen-specific cytotoxicity toward tumor cells in vitro. Mechanistically, loss of DOT1L resulted in an altered cell state with loss of T cell and gain of innate-like features. These transcriptional changes were mediated by loss of DOT1L methyltransferase activity in a dose-dependent manner. Our findings show that in mature CD8 T cells, ablation of DOT1L activity is well tolerated and reprograms them to gain innate-like memory cell characteristics and enhance intrinsic cytotoxic capacity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Kwesi-Maliepaard E. M., Aslam M. A., Alemdehy M. F., van den Brand T., McLean C., Vlaming H., van Welsem T., Korthout T., Lancini C., Hendriks S., Ahrends T., van Dinther D., den Haan J. M. M., Borst J., de Wit E., van Leeuwen F., Jacobs H., The histone methyltransferase DOT1L prevents antigen-independent differentiation and safeguards epigenetic identity of CD8+ T cells. Proc. Natl. Acad. Sci. U.S.A. 117, 20706–20716 (2020). - PMC - PubMed
-
- Vlaming H., van Leeuwen F., The upstreams and downstreams of H3K79 methylation by DOT1L. Chromosoma 125, 593–605 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

