Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes
- PMID: 6958982
- PMCID: PMC4881848
- DOI: 10.1038/300611a0
Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes
Abstract
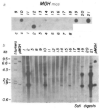
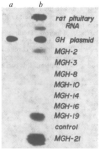
A DNA fragment containing the promoter of the mouse metallothionein-I gene fused to the structural gene of rat growth hormone was microinjected into the pronuclei of fertilized mouse eggs. Of 21 mice that developed from these eggs, seven carried the fusion gene and six of these grew significantly larger than their littermates. Several of these transgenic mice had extraordinarily high levels of the fusion mRNA in their liver and growth hormone in their serum. This approach has implications for studying the biological effects of growth hormone, as a way to accelerate animal growth, as a model for gigantism, as a means of correcting genetic disease, and as a method of farming valuable gene products.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

