Dual function transcripts specifying tRNA and mRNA
- PMID: 7312036
- PMCID: PMC1987712
- DOI: 10.1038/294422a0
Dual function transcripts specifying tRNA and mRNA
Abstract
A cluster of four tRNA genes in Escherichia coli is co-transcribed with an adjacent gene encoding elongation factor Tu. The resultant transcript that specifies both structural (tRNA) and informational (mRNA) RNA may not be an uncommon occurrence and has interesting regulatory implications.
Figures

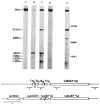

 ) is denoted by a + in the appropriate restriction fragment (from λrifd18) and lack of hybridization by −. The HinfI-82 probe from the
region hybridized strongly to 16S RNA in addition to the 1,800-base transcript. This result is consistent with the presence of a number of 12–16-bp homologies between 16S RNA and
sequences. Due to the hybridization with 16S RNA, the HinfI-82 probe could not be used to determine whether the smaller tuf transcripts (1,550 and 1,420 bases shown in Fig. 2) were tufA or tufB specific. The scale is in bases. Structural sequences for the tRNAs and tufB are boxed.
) is denoted by a + in the appropriate restriction fragment (from λrifd18) and lack of hybridization by −. The HinfI-82 probe from the
region hybridized strongly to 16S RNA in addition to the 1,800-base transcript. This result is consistent with the presence of a number of 12–16-bp homologies between 16S RNA and
sequences. Due to the hybridization with 16S RNA, the HinfI-82 probe could not be used to determine whether the smaller tuf transcripts (1,550 and 1,420 bases shown in Fig. 2) were tufA or tufB specific. The scale is in bases. Structural sequences for the tRNAs and tufB are boxed.
 , BstI; ↓, HaeIII; ↥, HhaI;
, BstI; ↓, HaeIII; ↥, HhaI;
 HincII;
HincII;
 HinfI;
HinfI;
 SmaI.
SmaI.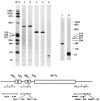

 ), processing intermediates and mature tRNAs (
), processing intermediates and mature tRNAs (
 ) are labelled by their respective sizes (in bases) on denaturing gels. The probe containing the structural tyrosine tRNA sequence (B) hybridized to a 230-base transcript that also hybridized with a
upstream probe and must therefore be a tyrT transcript. Probe C contained only 18 bases of structural tyrosine tRNA sequence, which severely limited its binding to the 200- or 230-base tyrosine transcripts in the stringent washing conditions used in these experiments. In the blot shown, the hybridization of probe C to the 170-base transcript did not photograph; in other blots this probe detectably bound the 170-base transcript. The processing scheme is discussed in the text.
) are labelled by their respective sizes (in bases) on denaturing gels. The probe containing the structural tyrosine tRNA sequence (B) hybridized to a 230-base transcript that also hybridized with a
upstream probe and must therefore be a tyrT transcript. Probe C contained only 18 bases of structural tyrosine tRNA sequence, which severely limited its binding to the 200- or 230-base tyrosine transcripts in the stringent washing conditions used in these experiments. In the blot shown, the hybridization of probe C to the 170-base transcript did not photograph; in other blots this probe detectably bound the 170-base transcript. The processing scheme is discussed in the text.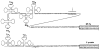
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

