A cluster of five nuclear proteins regulates keratin gene transcription
- PMID: 7505672
- PMCID: PMC6081630
A cluster of five nuclear proteins regulates keratin gene transcription
Abstract
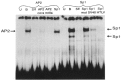
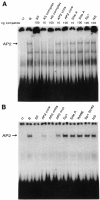
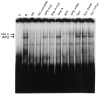
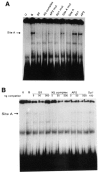
A common feature of all epithelial cells is the presence of keratin proteins that assemble into an intermediate filament cytoskeletal network. Whereas other cell types often use a specific master transcription factor to coordinate cell type-specific transcription, analysis of transcriptional regulation of keratin genes suggests that specific groupings of widely expressed transcription factors, acting on clusters of recognition elements in the promoter regions, confer epithelia-specific transcription. We define such a cluster of three sites that binds five transcription factors in the human K5 keratin gene. Within this cluster, an unusual Sp1 site binds the Sp1 transcription factor and two additional proteins. Flanking the Sp1 site are an AP2 site and another sequence, Site A; each binds a transcription factor. Similar clusters of recognition sites for the same five transcription factors have been also identified in other keratin genes. Such clusters may play a role in epithelia-specific expression of keratins.
Figures
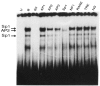
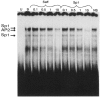
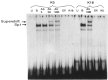
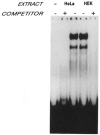
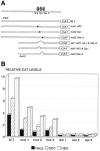

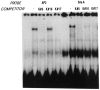

Similar articles
-
Tissue-specific in vivo protein-DNA interactions at the promoter region of the Xenopus 63 kDa keratin gene during metamorphosis.Nucleic Acids Res. 1995 Nov 11;23(21):4502-9. doi: 10.1093/nar/23.21.4502. Nucleic Acids Res. 1995. PMID: 7501476 Free PMC article.
-
Transcriptional regulators of expression of K#16, the disease-associated keratin.DNA Cell Biol. 1993 Dec;12(10):911-23. doi: 10.1089/dna.1993.12.911. DNA Cell Biol. 1993. PMID: 7506038
-
High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter.Virology. 1996 Oct 1;224(1):281-91. doi: 10.1006/viro.1996.0530. Virology. 1996. PMID: 8862423
-
Transcriptional regulation of the hepatocyte growth factor (HGF) gene by the Sp family of transcription factors.Oncogene. 1997 Jun 26;14(25):3039-49. doi: 10.1038/sj.onc.1201152. Oncogene. 1997. PMID: 9223667
-
Characterization of nuclear protein binding sites in the promoter of keratin K17 gene.DNA Cell Biol. 1996 Jan;15(1):65-74. doi: 10.1089/dna.1996.15.65. DNA Cell Biol. 1996. PMID: 8561898
Cited by
-
Novel mechanism of steroid action in skin through glucocorticoid receptor monomers.Mol Cell Biol. 2000 Jun;20(12):4328-39. doi: 10.1128/MCB.20.12.4328-4339.2000. Mol Cell Biol. 2000. PMID: 10825196 Free PMC article.
-
Disease-activated transcription factor: allergic reactions in human skin cause nuclear translocation of STAT-91 and induce synthesis of keratin K17.Mol Cell Biol. 1994 Jul;14(7):4759-69. doi: 10.1128/mcb.14.7.4759-4769.1994. Mol Cell Biol. 1994. PMID: 7516473 Free PMC article.
-
On the role of AP2 in epithelial-specific gene expression.Gene Expr. 1993;3(3):307-15. Gene Expr. 1993. PMID: 7517240 Free PMC article.
-
Transcriptional control of K5, K6, K14, and K17 keratin genes by AP-1 and NF-kappaB family members.Gene Expr. 1997;6(6):361-70. Gene Expr. 1997. PMID: 9495317 Free PMC article.
-
An Sp1 binding site and the minimal promoter contribute to overexpression of the cytokeratin 18 gene in tumorigenic clones relative to that in nontumorigenic clones of a human carcinoma cell line.Mol Cell Biol. 1995 May;15(5):2490-9. doi: 10.1128/MCB.15.5.2490. Mol Cell Biol. 1995. PMID: 7537848 Free PMC article.
References
-
- Briggs M. R., Kadonaga J. T., Bell S. P., and Tjian R. (1986), Science 234, 47–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
