The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment
- PMID: 7522904
- PMCID: PMC4167666
- DOI: 10.1016/s0969-2126(00)00062-9
The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment
Abstract
Background: Members of the Parvovirus genus cause a variety of diseases in mammals, including humans. One of the major defences against viral infection is the presence of neutralizing antibodies that prevent virus particles from infecting target cells. The mechanism of neutralization is not well understood. We therefore studied the structure of canine parvovirus (CPV) complexed with the Fab fragment of a neutralizing antibody, A3B10, using image reconstruction of electron micrographs of vitrified samples, together with the already known structure of CPV from X-ray crystallographic data.
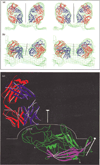
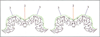
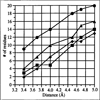
Results: The structure of the complex of CPV with Fab A3B10 has been determined to 23 A resolution. The known CPV atomic structure was subtracted from the electron density of the complex, and the difference map was used to fit the atomic coordinates of a known Fab fragment, HyHEL-5. The long axis of each Fab molecule is oriented in a near radial direction, inclined away from the two-fold axes. The viral epitope consists of 14 amino acid residues found in loops 1, 2 and 3 on the capsid surface, which include previously identified escape mutations.
Conclusions: The mode of Fab binding suggests that the A3B10 neutralizing antibody cannot bind bivalently to the capsid across the two-fold axes, consistent with the observation that whole A3B10 antibody readily precipitates CPV. Since Fab A3B10 can also neutralize the virus, mechanisms of neutralization such as interference with cell attachment, cell entry, or uncoating, must be operative.
Figures





Similar articles
-
Structural comparison of different antibodies interacting with parvovirus capsids.J Virol. 2009 Jun;83(11):5556-66. doi: 10.1128/JVI.02532-08. Epub 2009 Mar 25. J Virol. 2009. PMID: 19321620 Free PMC article.
-
Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody.J Virol. 1993 Mar;67(3):1148-58. doi: 10.1128/JVI.67.3.1148-1158.1993. J Virol. 1993. PMID: 7679742 Free PMC article.
-
Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.J Virol. 2016 Oct 14;90(21):9733-9742. doi: 10.1128/JVI.01112-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27535057 Free PMC article.
-
Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop.EMBO J. 1997 Apr 1;16(7):1492-500. doi: 10.1093/emboj/16.7.1492. EMBO J. 1997. PMID: 9130694 Free PMC article.
-
Viral Capsid, Antibody, and Receptor Interactions: Experimental Analysis of the Antibody Escape Evolution of Canine Parvovirus.J Virol. 2023 Jun 29;97(6):e0009023. doi: 10.1128/jvi.00090-23. Epub 2023 May 18. J Virol. 2023. PMID: 37199627 Free PMC article.
Cited by
-
Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid.J Virol. 1997 Dec;71(12):9214-22. doi: 10.1128/JVI.71.12.9214-9222.1997. J Virol. 1997. PMID: 9371580 Free PMC article.
-
Adeno-associated virus serotype 1 (AAV1)- and AAV5-antibody complex structures reveal evolutionary commonalities in parvovirus antigenic reactivity.J Virol. 2015 Feb;89(3):1794-808. doi: 10.1128/JVI.02710-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410874 Free PMC article.
-
Atomic Resolution Structure of the Oncolytic Parvovirus LuIII by Electron Microscopy and 3D Image Reconstruction.Viruses. 2017 Oct 30;9(11):321. doi: 10.3390/v9110321. Viruses. 2017. PMID: 29084163 Free PMC article.
-
Structural comparison of different antibodies interacting with parvovirus capsids.J Virol. 2009 Jun;83(11):5556-66. doi: 10.1128/JVI.02532-08. Epub 2009 Mar 25. J Virol. 2009. PMID: 19321620 Free PMC article.
-
Canine parvovirus type 2c in Vietnam continues to produce distinct descendants with new mutations restricted to Vietnamese variants.Arch Virol. 2021 Jun;166(6):1741-1749. doi: 10.1007/s00705-021-05059-1. Epub 2021 Apr 16. Arch Virol. 2021. PMID: 33860842
References
-
- Studdert MJ. Tissue tropism of parvoviruses. In: Tijssen P, editor. CRC Handbook of Parvoviruses. II. Boca Raton, FL: CRC Press, Inc; 1990. pp. 3–27.
-
- Anderson MJ, Pereira MS, et al. Human parvovirus, the cause of erythema infectiosum (fifth disease)? Lancet. 1983;i:1378. - PubMed
-
- Siegl G. Biology and pathogenicity of autonomous parvoviruses. In: Berns KI, editor. The Parvoviruses. New York & London: Plenum Press; 1984. pp. 297–362.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

