Inserting the Ftz homeodomain into engrailed creates a dominant transcriptional repressor that specifically turns off Ftz target genes in vivo
- PMID: 7600995
- PMCID: PMC2749471
- DOI: 10.1242/dev.121.6.1801
Inserting the Ftz homeodomain into engrailed creates a dominant transcriptional repressor that specifically turns off Ftz target genes in vivo
Abstract
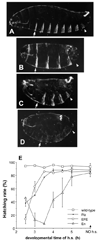
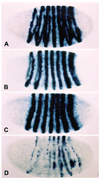
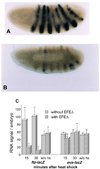
The Engrailed homeodomain protein is an 'active' or dominant transcriptional repressor in cultured cells. In contrast, the Fushi Tarazu homeodomain protein is an activator, both in cultured cells and in Drosophila embryos, where it activates several known target genes, including its own gene. This auto-activation has been shown to depend on targeting to a fushi tarazu enhancer by the Fushi Tarazu homeodomain. We combined Fushi Tarazu targeting and Engrailed active repression in a chimeric regulator, EFE. When EFE is ubiquitously expressed, it overrides endogenous Fushi Tarazu and causes a fushi tarazu mutant phenotype. Normal Fushi Tarazu target genes are affected as they are in fushi tarazu mutants. One such target gene is repressed by EFE even where Fushi Tarazu is not expressed, suggesting that the repression is active. This is confirmed by showing that the in vivo activity of EFE depends on a domain that is required for active repression in culture. A derivative that lacks this domain, while it cannot repress the endogenous fushi tarazu gene, can still reduce the activity of the fushi tarazu autoregulatory enhancer, suggesting that it competes with endogenous Fushi Tarazu for binding sites in vivo. However, this passive repression is much less effective than active repression.
Figures






References
-
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. - PubMed
-
- Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. - PubMed
-
- Baker N. Localization of transcripts from the wingless gene in whole Drosophila embryos. Development. 1988;103:289–298. - PubMed
-
- Biggin MD, Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989;58:433–440. - PubMed
-
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–312. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

