Localization of centromere function in a Drosophila minichromosome
- PMID: 7664339
- PMCID: PMC3209481
- DOI: 10.1016/0092-8674(95)90032-2
Localization of centromere function in a Drosophila minichromosome
Abstract
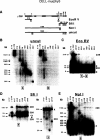
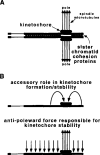
The DNA elements responsible for centromere activity in a metazoan have been localized using the Drosophila minichromosome Dp1187. Deleted minichromosomes were generated by irradiation mutagenesis, and their molecular structures were determined by pulsed-field Southern blot analysis. Analyses of the transmission behavior of Dp1187 derivatives localized sequences necessary for chromosome inheritance within the centric heterochromatin. The essential core of the centromere is contained within a 220 kb region that includes significant amounts of complex DNA. Completely normal inheritance also requires approximately 200 kb on either side of the essential core. This flanking DNA predominantly contains highly repeated sequences, and the amount required for normal transmission differs among division types and between the sexes. We propose that the essential core is the site of kinetochore formation and that flanking DNA provides two functions: sister chromatid cohesion and indirect assistance in kinetochore formation or function.
Figures





References
-
- Afshar K, Barton NR, Hawley RS, Goldstein LSB. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell. 1995;81:129–138. - PubMed
-
- Allshire RC, Cranston G, Gosden JR, Maule JC, Hastie ND, Fantes PA. A fission yeast chromosome can replicate autonomously in mouse cells. Cell. 1987;50:391–403. - PubMed
-
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. - PubMed
-
- Ashburner M. Drosophila: A Laboratory Handbook. Cold Spring Harbor Press; Cold Spring Harbor, New York: 1990.
-
- Ault JG, Rieder CL. Centrosome and kinetochore movement during mitosis. Curr. Opin. Cell Biol. 1994;6:41–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

