Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle
- PMID: 7720557
- PMCID: PMC2753456
- DOI: 10.1242/dev.120.11.3131
Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle
Abstract
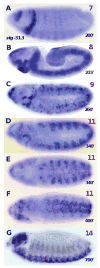
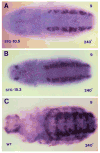
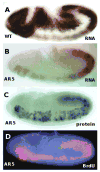
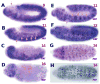
During postblastoderm embryogenesis in Drosophila, cell cycles progress in an invariant spatiotemporal pattern. Most of these cycles are differentially timed by bursts of transcription of string (cdc25), a gene encoding a phosphatase that triggers mitosis by activating the Cdc2 kinase. An analysis of string expression in 36 pattern-formation mutants shows that known patterning genes act locally to influence string transcription. Embryonic expression of string gene fragments shows that the complete pattern of string transcription requires extensive cis-acting regulatory sequences (> 15.3 kb), but that smaller segments of this regulatory region can drive proper temporal expression in defined spatial domains. We infer that string upstream sequences integrate many local signals to direct string's transcriptional program. Finally, we show that the spatiotemporal progression of string transcription is largely unaffected in mutant embryos specifically arrested in G2 of cycles 14, 15, or 16, or G1 of cycle 17. Thus, there is a regulatory hierarchy in which developmental inputs, not cell cycle inputs, control the timing of string transcription and hence cell cycle progression.
Figures



References
-
- Arora K, Nusslein-Volhard C. Altered mitotic domains reveal fate map changes in Drosophila embryos mutant for zygotic dorsoventral patterning genes. Development. 1992;114:1003–1024. - PubMed
-
- Ashburner M. Drosophila, a laboratory handbook. Cold Spring Harbor Press; 1989.
-
- Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; 1993.
-
- Bodmer R, Carretto R, Jan YN. Neurogenesis of the Peripheral Nervous System in Drosophila Embryos: DNA Replication Patterns and Cell Lineages. Neuron. 1989;3:21–32. - PubMed
-
- Boulay JL, Dennefeld C, Alberga A. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature. 1987;330:395–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

