A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells
- PMID: 8105392
- PMCID: PMC9524217
- DOI: 10.1038/365666a0
A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells
Abstract
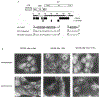
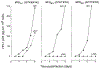
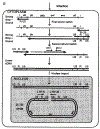
Permissiveness of the host cell to productive infection by oncoretroviruses is cell-cycle dependent, and nuclear localization of viral nucleoprotein preintegration complexes will occur only after cells have passed through mitosis. In contrast, establishment of an integrated provirus after infection by the lentivirus HIV-1 is independent of host cell proliferation. The ability of HIV-1 to replicate in non-dividing cells is partly accounted for by the karyophilic properties of the viral preintegration complex which, after virus infection, is actively transported to the host cell nucleus. Here we report that the gag matrix protein of HIV-1 contains a nuclear localization sequence which, when conjugated to a heterologous protein, directs its nuclear import. In addition, HIV-1 mutants containing amino-acid substitutions in this nuclear localization signal integrate and replicate within dividing but not growth-arrested cells, and thus display a phenotype more representative of an oncoretrovirus.
Figures


Comment in
-
HIV-1 infection of non-dividing cells.Nature. 1994 May 12;369(6476):107-8. doi: 10.1038/369107b0. Nature. 1994. PMID: 8192816 No abstract available.
References
-
- Varmus HE & Swanstrom R in RNA Tumor Viruses (eds Weiss R, Teich N, Varmus HE & Coffin J) 369–512 (Cold Spring Harbor Laboratory Press, New York, 1984).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

