Molecular organization of the rat glia-derived nexin/protease nexin-1 promoter
- PMID: 8268720
- PMCID: PMC6081634
Molecular organization of the rat glia-derived nexin/protease nexin-1 promoter
Abstract
The first three exons and the promoter of rat glia-derived nexin, also called protease nexin-1 (GDN/PN-1), have been identified through analysis of rat genomic clones. A 1.6 kilobase (kb) fragment containing 105 base pairs of the first exon and 5'-flanking sequences was sequenced. The 5'-flanking sequence and the first exon were found to be GC-rich, indicating that the 5' region of the rat GDN/PN-1 gene resides within a CpG island. A TATA box-like sequence, but no CAAT box, was found. The rat GDN/PN-1 promoter contains five SP1 consensus sites, four consensus sites for the MyoD1 transcription factor, and one binding site for the transcription factors NGFI-A, NGFI-C, Krox-20, and Wilms tumor factor. The presence of these consensus sequences is consistent with the known expression pattern of GDN/PN-1. Primer extension and RNase protection assays identified one transcriptional start site. The 1.6 kb promoter fragment cloned in a reporter plasmid was found to induce firefly luciferase expression in a cell-specific manner. A positive regulatory element is localized in the region -1545 to -389. In vitro CpG methylation blocked transcription from the GDN/PN-1 promoter in rat hepatoma cells but not in C6 rat glioma cells.
Figures


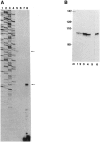


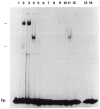
References
-
- Adams R. L. P., Bryans M., Rinaldi A., and Yesufu H. M. I. (1990), Philos Trans R Soc Lond Biol 326, 189–198. - PubMed
-
- Artelt P., Grannemann R., Stocking C., Friel J., Bartsch J., and Hauser H. (1991), Gene 99, 249–254. - PubMed
-
- Beato M. (1989), Cell 56, 335–344. - PubMed
-
- Bird A.P. (1986), Nature 321, 209–213. - PubMed
-
- Boyes J. and Bird A. P. (1991), Cell 64, 1123–1134. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous
