The two functionally distinct amino termini of chicken c-ets-1 products arise from alternative promoter usage
- PMID: 8268721
- PMCID: PMC6081633
The two functionally distinct amino termini of chicken c-ets-1 products arise from alternative promoter usage
Abstract
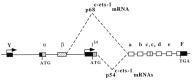
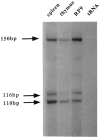
The chicken c-ets-1 locus gives rise to two distinct transcription factors differing by structurally and functionally unrelated N-termini. p54c-ets-1 shows a striking phylogenetic conservation from Xenopus to humans, while p68c-ets-1, the cellular counterpart of the E26-derived v-ets oncogene, is apparently restricted to avian and reptilian species. In the chick embryo, both mRNAs are expressed in a wide array of tissues of mesodermal origin; however, in the embryo and after hatching, p68c-ets-1 is excluded from lymphoid cells where p54c-ets-1 accumulates. In this report, we define the basis of the differential expression of the chicken c-ets-1 products to assess their different potentials as transcription factors. We demonstrate that the two distinct N-termini arise from alternative promoter usage within the chicken c-ets-1 locus. Examination of both promoters reveals that transcription initiates from multiple sites, consistent with the absence of TATA and CAAT elements. Of these two regulatory regions, only the one that initiates the p54c-ets-1 mRNA synthesis is of the G + C-rich type, and its organization is conserved in humans. The avian-specific p68c-ets-1 promoter activity was enhanced by its own product. In addition, we identify numerous potential binding sites for lymphoid-specific transcription factors that might contribute to a tight repressor effect in lymphoid tissues.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
