Dual roles of plcA in Listeria monocytogenes pathogenesis
- PMID: 8388529
- PMCID: PMC4836944
- DOI: 10.1111/j.1365-2958.1993.tb01211.x
Dual roles of plcA in Listeria monocytogenes pathogenesis
Abstract
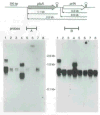
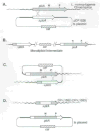
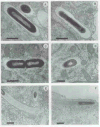
The plcA gene of Listeria monocytogenes encodes a secreted phosphatidylinositol-specific phospholipase C (Pl-PLC). Recent studies have established that transposon mutations within plcA result in avirulence for mice and pleiotropic effects when examined in tissue-culture models of infection. Genetic analysis reveals that many of the effects of the transposon insertions are due to loss of readthrough transcription from plcA into the downstream gene prfA, which encodes an essential transcription factor of numerous L. monocytogenes virulence genes. Construction of an in-frame deletion within plcA had no effect on expression of prfA thus allowing direct assignment of a role of the Pl-PLC in pathogenesis. Pl-PLC was shown to play a significant role in mediating escape of L. monocytogenes from phagosomes of primary murine macrophages. Interestingly, this defect manifested itself in vivo in the liver but not in the spleen of infected mice.
Figures







References
-
- Behnke D, Gilmore MS. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mot Gen Genet. 1981;184:115–120. - PubMed
-
- Breton CB, Langsley G, Barale J, Pereira da Silva LH. A malaria phosphatidylinositol-specific phospholipase C: a possible role in merozoite maturation and erythrocyte invasion. In: Cohen CM, Palek J, editors. Cellular and Molecular Biology of Normal and Abnormal Erythroid Membranes. New York: Wiley-Liss; 1990. pp. 315–332.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

